IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage
- PMID: 24065129
- PMCID: PMC3801437
- DOI: 10.1038/emboj.2013.203
IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage
Abstract
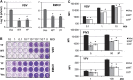
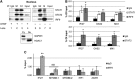
A single high dose of interferon-β (IFNβ) activates powerful cellular responses, in which many anti-viral, pro-apoptotic, and anti-proliferative proteins are highly expressed. Since some of these proteins are deleterious, cells downregulate this initial response rapidly. However, the expression of many anti-viral proteins that do no harm is sustained, prolonging a substantial part of the initial anti-viral response for days and also providing resistance to DNA damage. While the transcription factor ISGF3 (IRF9 and tyrosine-phosphorylated STATs 1 and 2) drives the first rapid response phase, the related factor un-phosphorylated ISGF3 (U-ISGF3), formed by IFNβ-induced high levels of IRF9 and STATs 1 and 2 without tyrosine phosphorylation, drives the second prolonged response. The U-ISGF3-induced anti-viral genes that show prolonged expression are driven by distinct IFN stimulated response elements (ISREs). Continuous exposure of cells to a low level of IFNβ, often seen in cancers, leads to steady-state increased expression of only the U-ISGF3-dependent proteins, with no sustained increase in other IFNβ-induced proteins, and to constitutive resistance to DNA damage.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Andersen JB, Hassel BA (2006) The interferon regulated ubiquitin-like protein, ISG15, in tumorigenesis: friend or foe? Cytokine Growth Factor Rev 17: 411–42a - PubMed
-
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

