In silico modeling of Itk activation kinetics in thymocytes suggests competing positive and negative IP4 mediated feedbacks increase robustness
- PMID: 24066087
- PMCID: PMC3774804
- DOI: 10.1371/journal.pone.0073937
In silico modeling of Itk activation kinetics in thymocytes suggests competing positive and negative IP4 mediated feedbacks increase robustness
Erratum in
- PLoS One. 2014;9(1): 10.1371/annotation/285bb79f-ef1a-467e-9fd3-4e518d65acd3
Abstract
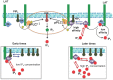
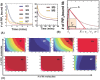
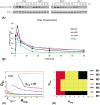
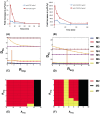
The inositol-phosphate messenger inositol(1,3,4,5)tetrakisphosphate (IP4) is essential for thymocyte positive selection by regulating plasma-membrane association of the protein tyrosine kinase Itk downstream of the T cell receptor (TCR). IP4 can act as a soluble analog of the phosphoinositide 3-kinase (PI3K) membrane lipid product phosphatidylinositol(3,4,5)trisphosphate (PIP3). PIP3 recruits signaling proteins such as Itk to cellular membranes by binding to PH and other domains. In thymocytes, low-dose IP4 binding to the Itk PH domain surprisingly promoted and high-dose IP4 inhibited PIP3 binding of Itk PH domains. However, the mechanisms that underlie the regulation of membrane recruitment of Itk by IP4 and PIP3 remain unclear. The distinct Itk PH domain ability to oligomerize is consistent with a cooperative-allosteric mode of IP4 action. However, other possibilities cannot be ruled out due to difficulties in quantitatively measuring the interactions between Itk, IP4 and PIP3, and in generating non-oligomerizing Itk PH domain mutants. This has hindered a full mechanistic understanding of how IP4 controls Itk function. By combining experimentally measured kinetics of PLCγ1 phosphorylation by Itk with in silico modeling of multiple Itk signaling circuits and a maximum entropy (MaxEnt) based computational approach, we show that those in silico models which are most robust against variations of protein and lipid expression levels and kinetic rates at the single cell level share a cooperative-allosteric mode of Itk regulation by IP4 involving oligomeric Itk PH domains at the plasma membrane. This identifies MaxEnt as an excellent tool for quantifying robustness for complex TCR signaling circuits and provides testable predictions to further elucidate a controversial mechanism of PIP3 signaling.
Conflict of interest statement
Figures
References
-
- Irvine RF, Schell MJ (2001) Back in the water: the return of the inositol phosphates. Nature reviews Molecular cell biology 2: 327–338. - PubMed
-
- Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, et al. (2007) Positive regulation of Itk PH domain function by soluble IP4. Science 316: 886–889. - PubMed
-
- York JD (2006) Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta 1761: 552–559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

