C-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: analysis of the reaction mechanism and novel roles for pGpG
- PMID: 24066157
- PMCID: PMC3774798
- DOI: 10.1371/journal.pone.0074920
C-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: analysis of the reaction mechanism and novel roles for pGpG
Abstract
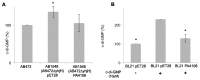
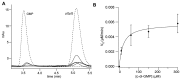
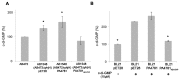
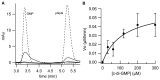
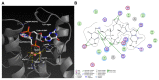
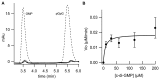
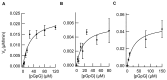
In biofilms, the bacterial community optimizes the strategies to sense the environment and to communicate from cell to cell. A key player in the development of a bacterial biofilm is the second messenger c-di-GMP, whose intracellular levels are modulated by the opposite activity of diguanylate cyclases and phosphodiesterases. Given the huge impact of bacterial biofilms on human health, understanding the molecular details of c-di-GMP metabolism represents a critical step in the development of novel therapeutic approaches against biofilms. In this study, we present a detailed biochemical characterization of two c-di-GMP phosphodiesterases of the HD-GYP subtype from the human pathogen Pseudomonas aeruginosa, namely PA4781 and PA4108. Upstream of the catalytic HD-GYP domain, PA4781 contains a REC domain typical of two-component systems, while PA4108 contains an uncharacterized domain of unknown function. Our findings shed light on the activity and catalytic mechanism of these phosphodiesterases. We show that both enzymes hydrolyse c-di-GMP in a two-step reaction via the linear intermediate pGpG and that they produce GMP in vitro at a surprisingly low rate. In addition, our data indicate that the non-phosphorylated REC domain of PA4781 prevents accessibility of c-di-GMP to the active site. Both PA4108 and phosphorylated PA4781 are also capable to use pGpG as an alternative substrate and to hydrolyse it into GMP; the affinity of PA4781 for pGpG is one order of magnitude higher than that for c-di-GMP. These results suggest that these enzymes may not work (primarily) as genuine phosphodiesterases. Moreover, the unexpected affinity of PA4781 for pGpG may indicate that pGpG could also act as a signal molecule in its own right, thus further widening the c-di-GMP-related signalling scenario.
Conflict of interest statement
Figures
References
-
- Römling U, Gomelsky M, Galperin MY (2005) C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57: 629-639. doi:10.1111/j.1365-2958.2005.04697.x. PubMed: 16045609. - DOI - PubMed
-
- Schirmer T, Jenal U (2009) Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7: 724-735. doi:10.1038/nrmicro2203. PubMed: 19756011. - DOI - PubMed
-
- Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J et al. (2006) Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3'-5')-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103: 2839-2844. doi:10.1073/pnas.0511090103. PubMed: 16477007. - DOI - PMC - PubMed
-
- Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Barthel M et al. (2011) Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLOS ONE 6: e28351. doi:10.1371/journal.pone.0028351. PubMed: 22164276. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

