Systematic analyses of the cytotoxic effects of compound 11a, a putative synthetic agonist of photoreceptor-specific nuclear receptor (PNR), in cancer cell lines
- PMID: 24066170
- PMCID: PMC3774666
- DOI: 10.1371/journal.pone.0075198
Systematic analyses of the cytotoxic effects of compound 11a, a putative synthetic agonist of photoreceptor-specific nuclear receptor (PNR), in cancer cell lines
Abstract
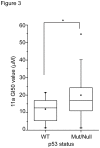
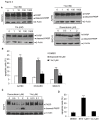
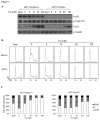
Photoreceptor cell-specific receptor (PNR/NR2E3) is an orphan nuclear receptor that plays a critical role in retinal development and photoreceptor maintenance. The disease-causing mutations in PNR have a pleiotropic effect resulting in varying retinal diseases. Recently, PNR has been implicated in control of cellular functions in cancer cells. PNR was reported to be a novel regulator of ERα expression in breast cancer cells, and high PNR expression correlates with favorable response to tamoxifen treatment. Moreover, PNR was shown to increase p53 stability in HeLa cells, implying that PNR may be a therapeutic target in this and other cancers that retain a wild type p53 gene. To facilitate further understanding of PNR functions in cancer, we characterized compound 11a, a synthetic, putative PNR agonist in several cell-based assays. Interestingly, we showed that 11a failed to activate PNR and its cytotoxicity was independent of PNR expression, excluding PNR as a mediator for 11a cytotoxicity. Systematic analyses of the cytotoxic effects of 11a in NCI-60 cell lines revealed a strong positive correlation of cytotoxicity with p53 status, i.e., p53 wild type cell lines were significantly more sensitive to 11a than p53 mutated or null cell lines. Furthermore, using HCT116 p53+/+ and p53-/- isogenic cell lines we revealed that the mechanism of 11a-induced cytotoxicity occurred through G1/S phase cell cycle arrest rather than apoptosis. In conclusion, we observed a correlation of 11a sensitivity with p53 status but not with PNR expression, suggesting that tumors expressing wild type p53 might be responsive to this compound.
Conflict of interest statement
Figures




Similar articles
-
Orphan nuclear receptor NR2E3 is a new molecular vulnerability in solid tumors by activating p53.Cell Death Dis. 2025 Jan 14;16(1):15. doi: 10.1038/s41419-025-07337-1. Cell Death Dis. 2025. PMID: 39809731 Free PMC article.
-
Orphan nuclear receptor PNR/NR2E3 stimulates p53 functions by enhancing p53 acetylation.Mol Cell Biol. 2012 Jan;32(1):26-35. doi: 10.1128/MCB.05513-11. Epub 2011 Oct 24. Mol Cell Biol. 2012. PMID: 22025681 Free PMC article.
-
In pursuit of synthetic modulators for the orphan retina-specific nuclear receptor NR2E3.J Ocul Pharmacol Ther. 2013 Apr;29(3):298-309. doi: 10.1089/jop.2012.0135. Epub 2012 Oct 25. J Ocul Pharmacol Ther. 2013. PMID: 23098562 Free PMC article.
-
Heterodimers of photoreceptor-specific nuclear receptor (PNR/NR2E3) and peroxisome proliferator-activated receptor-γ (PPARγ) are disrupted by retinal disease-associated mutations.Cell Death Dis. 2017 Mar 16;8(3):e2677. doi: 10.1038/cddis.2017.98. Cell Death Dis. 2017. PMID: 28300834 Free PMC article.
-
Investigation of the role of Bax, p21/Waf1 and p53 as determinants of cellular responses in HCT116 colorectal cancer cells exposed to the novel cytotoxic ruthenium(II) organometallic agent, RM175.Cancer Chemother Pharmacol. 2005 Jun;55(6):577-83. doi: 10.1007/s00280-004-0932-9. Epub 2005 Feb 22. Cancer Chemother Pharmacol. 2005. PMID: 15726367
Cited by
-
Somatic mutations of MLL4/COMPASS induce cytoplasmic localization providing molecular insight into cancer prognosis and treatment.Proc Natl Acad Sci U S A. 2023 Dec 26;120(52):e2310063120. doi: 10.1073/pnas.2310063120. Epub 2023 Dec 19. Proc Natl Acad Sci U S A. 2023. PMID: 38113256 Free PMC article.
-
Potential of Small Molecule-Mediated Reprogramming of Rod Photoreceptors to Treat Retinitis Pigmentosa.Invest Ophthalmol Vis Sci. 2016 Nov 1;57(14):6407-6415. doi: 10.1167/iovs.16-20177. Invest Ophthalmol Vis Sci. 2016. PMID: 27893103 Free PMC article.
-
Orphan nuclear receptor NR2E3 is a new molecular vulnerability in solid tumors by activating p53.Cell Death Dis. 2025 Jan 14;16(1):15. doi: 10.1038/s41419-025-07337-1. Cell Death Dis. 2025. PMID: 39809731 Free PMC article.
-
IL-13Rα2 mediates PNR-induced migration and metastasis in ERα-negative breast cancer.Oncogene. 2015 Mar 19;34(12):1596-607. doi: 10.1038/onc.2014.53. Epub 2014 Apr 21. Oncogene. 2015. PMID: 24747967 Free PMC article.
-
Therapeutic targeting of metabolic vulnerabilities in cancers with MLL3/4-COMPASS epigenetic regulator mutations.J Clin Invest. 2023 Jul 3;133(13):e169993. doi: 10.1172/JCI169993. J Clin Invest. 2023. PMID: 37252797 Free PMC article.
References
-
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ (2001) Nuclear receptors and lipid physiology: opening the X-files. Science 294: 1866-1870. doi:10.1126/science.294.5548.1866. PubMed: 11729302. - DOI - PubMed
-
- Francis GA, Fayard E, Picard F, Auwerx J (2003) Nuclear receptors and the control of metabolism. Annu Rev Physiol 65: 261-311. doi:10.1146/annurev.physiol.65.092101.142528. PubMed: 12518001. - DOI - PubMed
-
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G et al. (1995) The nuclear receptor superfamily: the second decade. Cell 83: 835-839. doi:10.1016/0092-8674(95)90199-X. PubMed: 8521507. - DOI - PMC - PubMed
-
- Tenbaum S, Baniahmad A (1997) Nuclear receptors: structure, function and involvement in disease. Int J Biochem Cell Biol 29: 1325-1341. doi:10.1016/S1357-2725(97)00087-3. PubMed: 9570131. - DOI - PubMed
-
- Sonoda J, Pei L, Evans RM (2008) Nuclear receptors: decoding metabolic disease. FEBS Lett 582: 2-9. doi:10.1016/j.febslet.2007.11.016. PubMed: 18023286. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

