Diverse sources of C. difficile infection identified on whole-genome sequencing
- PMID: 24066741
- PMCID: PMC3868928
- DOI: 10.1056/NEJMoa1216064
Diverse sources of C. difficile infection identified on whole-genome sequencing
Abstract
Background: It has been thought that Clostridium difficile infection is transmitted predominantly within health care settings. However, endemic spread has hampered identification of precise sources of infection and the assessment of the efficacy of interventions.
Methods: From September 2007 through March 2011, we performed whole-genome sequencing on isolates obtained from all symptomatic patients with C. difficile infection identified in health care settings or in the community in Oxfordshire, United Kingdom. We compared single-nucleotide variants (SNVs) between the isolates, using C. difficile evolution rates estimated on the basis of the first and last samples obtained from each of 145 patients, with 0 to 2 SNVs expected between transmitted isolates obtained less than 124 days apart, on the basis of a 95% prediction interval. We then identified plausible epidemiologic links among genetically related cases from data on hospital admissions and community location.
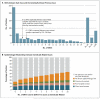
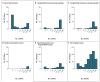
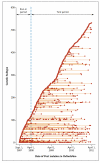
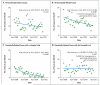
Results: Of 1250 C. difficile cases that were evaluated, 1223 (98%) were successfully sequenced. In a comparison of 957 samples obtained from April 2008 through March 2011 with those obtained from September 2007 onward, a total of 333 isolates (35%) had no more than 2 SNVs from at least 1 earlier case, and 428 isolates (45%) had more than 10 SNVs from all previous cases. Reductions in incidence over time were similar in the two groups, a finding that suggests an effect of interventions targeting the transition from exposure to disease. Of the 333 patients with no more than 2 SNVs (consistent with transmission), 126 patients (38%) had close hospital contact with another patient, and 120 patients (36%) had no hospital or community contact with another patient. Distinct subtypes of infection continued to be identified throughout the study, which suggests a considerable reservoir of C. difficile.
Conclusions: Over a 3-year period, 45% of C. difficile cases in Oxfordshire were genetically distinct from all previous cases. Genetically diverse sources, in addition to symptomatic patients, play a major part in C. difficile transmission. (Funded by the U.K. Clinical Research Collaboration Translational Infection Research Initiative and others.).
Figures
Comment in
-
Clostridium difficile--beyond the usual suspects.N Engl J Med. 2013 Sep 26;369(13):1263-4. doi: 10.1056/NEJMe1310454. N Engl J Med. 2013. PMID: 24066748 No abstract available.
-
What's your subtype? The epidemiologic utility of bacterial whole-genome sequencing.Clin Chem. 2014 Apr;60(4):586-8. doi: 10.1373/clinchem.2013.217141. Epub 2014 Jan 7. Clin Chem. 2014. PMID: 24398073 Free PMC article. No abstract available.
-
Diverse sources of C. difficile infection.N Engl J Med. 2014 Jan 9;370(2):183-4. doi: 10.1056/NEJMc1313601. N Engl J Med. 2014. PMID: 24401066 No abstract available.
-
Diverse sources of C. difficile infection.N Engl J Med. 2014 Jan 9;370(2):182. doi: 10.1056/NEJMc1313601. N Engl J Med. 2014. PMID: 24401067 No abstract available.
-
Diverse sources of C. difficile infection.N Engl J Med. 2014 Jan 9;370(2):182-3. doi: 10.1056/NEJMc1313601. N Engl J Med. 2014. PMID: 24401068 No abstract available.
References
-
- Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. - PubMed
-
- Vonberg R-P, Kuijper EJ, Wilcox MH, et al. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect. 2008;14(Suppl 5):2–20. - PubMed
-
- McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–10. - PubMed
-
- Samore MH, DeGirolami PC, Tlucko A, Lichtenberg DA, Melvin ZA, Karchmer AW. Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis. 1994;18:181–7. - PubMed
-
- Dubberke ER, Reske KA, Olsen MA, et al. Evaluation of Clostridium difficile-associated disease pressure as a risk factor for C. difficile-associated disease. Arch Intern Med. 2007;167:1092–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
