HIC1 interacts with and modulates the activity of STAT3
- PMID: 24067369
- PMCID: PMC3755077
- DOI: 10.4161/cc.25365
HIC1 interacts with and modulates the activity of STAT3
Abstract
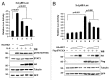
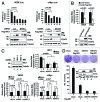
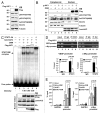
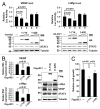
HIC1 (hypermethylated in cancer 1) is a tumor suppressor gene, expression of which is frequently suppressed in human cancers. Very little is known about the molecular basis of HIC1 in antagonizing oncogenic pathways. Here, we report that HIC1 forms complexes with the signal transducers and activators of transcription 3 (STAT3) and attenuates STAT3-mediated transcription. STAT3 was identified as a HIC1-interacting protein by affinity capture and followed by mass spectrometry analysis. Overexpression or depletion of HIC1 resulted in decreased or increased levels of interleukin-6 (IL-6)/oncostatin M (OSM)-induced STAT3-mediated reporter activity and expression of target genes such as VEGF and c-Myc, respectively. Furthermore, HIC1 suppressing the VEGF and c-Myc promoter activity and the colony formation of MDA-MB 231 cells were STAT3-dependent. Further studies showed that HIC1 interacts with the DNA binding domain of STAT3 and suppresses the binding of STAT3 to its target gene promoters. Domain mapping study revealed that HIC1 C-terminal domain binds to STAT3. HIC1 mutant defective in STAT3 interaction reduced its repressive effect on STAT3 DNA binding activity, the reporter activity and gene expression of the VEGF and c-Myc genes, and cell growth in MDA-MB 231 cells. Altogether, our findings not only provide a novel role of HIC1 in antagonizing STAT3-mediated activation of VEGF and c-Myc gene expression and cell growth, but also elucidate a molecular basis underlying the inhibitory effect of HIC1 on STAT3 transcriptional potential.
Keywords: DNA binding; HIC1; STAT3; protein interaction; transcriptional regulation.
Figures


Similar articles
-
HIC1 attenuates invasion and metastasis by inhibiting the IL-6/STAT3 signalling pathway in human pancreatic cancer.Cancer Lett. 2016 Jul 1;376(2):387-98. doi: 10.1016/j.canlet.2016.04.013. Epub 2016 Apr 13. Cancer Lett. 2016. PMID: 27085461
-
Hypermethylated in cancer 1(HIC1) suppresses non-small cell lung cancer progression by targeting interleukin-6/Stat3 pathway.Oncotarget. 2016 May 24;7(21):30350-64. doi: 10.18632/oncotarget.8734. Oncotarget. 2016. PMID: 27107418 Free PMC article.
-
Oncostatin M promotes STAT3 activation, VEGF production, and invasion in osteosarcoma cell lines.BMC Cancer. 2011 Apr 11;11:125. doi: 10.1186/1471-2407-11-125. BMC Cancer. 2011. PMID: 21481226 Free PMC article.
-
Implication of HIC1 (Hypermethylated In Cancer 1) in the DNA damage response.Bull Cancer. 2009 Nov;96(11):E66-72. doi: 10.1684/bdc.2009.0959. Bull Cancer. 2009. PMID: 19822477 Review.
-
HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors.Int J Biochem Cell Biol. 2009 Jan;41(1):26-33. doi: 10.1016/j.biocel.2008.05.028. Epub 2008 Aug 3. Int J Biochem Cell Biol. 2009. PMID: 18723112 Free PMC article. Review.
Cited by
-
Interleukin-6 serves as a critical factor in various cancer progression and therapy.Med Oncol. 2024 Jun 20;41(7):182. doi: 10.1007/s12032-024-02422-5. Med Oncol. 2024. PMID: 38900329 Review.
-
Genistein inhibits angiogenesis developed during rheumatoid arthritis through the IL-6/JAK2/STAT3/VEGF signalling pathway.J Orthop Translat. 2019 Aug 23;22:92-100. doi: 10.1016/j.jot.2019.07.007. eCollection 2020 May. J Orthop Translat. 2019. PMID: 32440504 Free PMC article.
-
The transcriptional repressor HIC1 regulates intestinal immune homeostasis.Mucosal Immunol. 2017 Nov;10(6):1518-1528. doi: 10.1038/mi.2017.17. Epub 2017 Mar 22. Mucosal Immunol. 2017. PMID: 28327618
-
STAT3 Interactors as Potential Therapeutic Targets for Cancer Treatment.Int J Mol Sci. 2018 Jun 16;19(6):1787. doi: 10.3390/ijms19061787. Int J Mol Sci. 2018. PMID: 29914167 Free PMC article. Review.
-
Pomegranate-specific natural compounds as onco-preventive and onco-therapeutic compounds: Comparison with conventional drugs acting on the same molecular mechanisms.Heliyon. 2023 Jul 8;9(7):e18090. doi: 10.1016/j.heliyon.2023.e18090. eCollection 2023 Jul. Heliyon. 2023. PMID: 37519687 Free PMC article. Review.
References
-
- Boulay G, Dubuissez M, Van Rechem C, Forget A, Helin K, Ayrault O, et al. Hypermethylated in cancer 1 (HIC1) recruits polycomb repressive complex 2 (PRC2) to a subset of its target genes through interaction with human polycomb-like (hPCL) proteins. J Biol Chem. 2012;287:10509–24. doi: 10.1074/jbc.M111.320234. - DOI - PMC - PubMed
-
- Van Rechem C, Boulay G, Pinte S, Stankovic-Valentin N, Guérardel C, Leprince D. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol. 2010;30:4045–59. doi: 10.1128/MCB.00582-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
