Discovery of a unique novel clade of mosquito-associated bunyaviruses
- PMID: 24067954
- PMCID: PMC3838119
- DOI: 10.1128/JVI.01862-13
Discovery of a unique novel clade of mosquito-associated bunyaviruses
Abstract
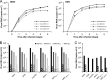
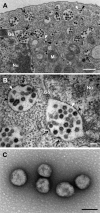
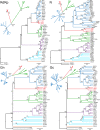
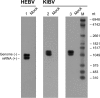
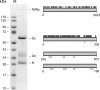
Bunyaviruses are the largest known family of RNA viruses, infecting vertebrates, insects, and plants. Here we isolated three novel bunyaviruses from mosquitoes sampled in Côte d'Ivoire, Ghana, and Uganda. The viruses define a highly diversified monophyletic sister clade to all members of the genus Orthobunyavirus and are virtually equidistant to orthobunyaviruses and tospoviruses. Maximal amino acid identities between homologous putative proteins of the novel group and orthobunyaviruses ranged between 12 and 25%. The type isolates, tentatively named Herbert virus (HEBV), Taï virus (TAIV), and Kibale virus (KIBV), comprised genomes with L, M, and S segments of about 7.4 kb, 2.7 kb, and 1.1 kb, respectively. HEBV, TAIV, and KIBV encode the shortest bunyavirus M segments known and did not seem to encode NSs and NSm proteins but contained an elongated L segment with an ∼500-nucleotide (nt) insertion that shows no identity to other bunyaviruses. The viruses replicated to high titers in insect cells but did not replicate in vertebrate cells. The enveloped virions were 90 to 110 nm in diameter and budded at cellular membranes with morphological features typical of the Golgi complex. Viral RNA recovered from infected cells showed 5'-terminal nontemplated sequences of 9 to 22 nt, suggestive of cap snatching during mRNA synthesis, as described for other bunyaviruses. Northern blotting identified RNA species of full and reduced lengths, suggested upon analogy with other bunyaviruses to constitute antigenomic-sense cRNA and transcript mRNAs, respectively. Functional studies will be necessary to determine if this group of viruses constitutes a novel genus in the bunyavirus family.
Figures




References
-
- Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB. 2012. Family Bunyaviridae, p 725–741 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus Taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA
-
- Bird BH, Nichol ST. 2012. Breaking the chain: Rift Valley fever virus control via livestock vaccination. Curr. Opin. Virol. 2:315–323 - PubMed
-
- Ergonul O. 2012. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr. Opin. Virol. 2:215–220 - PubMed
-
- Soldan SS, Gonzalez-Scarano F. 2005. Emerging infectious diseases: the Bunyaviridae. J. Neurovirol. 11:412–423 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

