A deubiquitinase negatively regulates retro-translocation of nonubiquitinated substrates
- PMID: 24068323
- PMCID: PMC3826992
- DOI: 10.1091/mbc.E13-06-0332
A deubiquitinase negatively regulates retro-translocation of nonubiquitinated substrates
Abstract
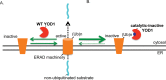
Endoplasmic reticulum (ER) membrane-bound E3 ubiquitin ligases promote ER-associated degradation (ERAD) by ubiquitinating a retro-translocated substrate that reaches the cytosol from the ER, targeting it to the proteasome for destruction. Recent findings implicate ERAD-associated deubiquitinases (DUBs) as positive and negative regulators during ERAD, reflecting the different consequences of deubiquitinating a substrate prior to proteasomal degradation. These observations raise the question of whether a DUB can control the fate of a nonubiquitinated ERAD substrate. In this study, we probed the role of the ERAD-associated DUB, YOD1, during retro-translocation of the nonubiquitinated cholera toxin A1 (CTA1) peptide, a critical intoxication step. Through combining knockdown, overexpression, and binding studies, we demonstrated that YOD1 negatively controls CTA1 retro-translocation, likely by deubiquitinating and inactivating ubiquitinated ERAD components that normally promote toxin retro-translocation. YOD1 also antagonizes the proteasomal degradation of nonglycosylated pro-α factor, a postulated nonubiquitinated yeast ERAD substrate, in mammalian cells. Our findings reveal that a cytosolic DUB exerts a negative function during retro-translocation of nonubiquitinated substrates, potentially by acting on elements of the ERAD machinery.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

