Using biological pathway data with paxtools
- PMID: 24068901
- PMCID: PMC3777916
- DOI: 10.1371/journal.pcbi.1003194
Using biological pathway data with paxtools
Abstract
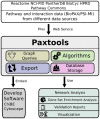
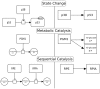
A rapidly growing corpus of formal, computable pathway information can be used to answer important biological questions including finding non-trivial connections between cellular processes, identifying significantly altered portions of the cellular network in a disease state and building predictive models that can be used for precision medicine. Due to its complexity and fragmented nature, however, working with pathway data is still difficult. We present Paxtools, a Java library that contains algorithms, software components and converters for biological pathways represented in the standard BioPAX language. Paxtools allows scientists to focus on their scientific problem by removing technical barriers to access and analyse pathway information. Paxtools can run on any platform that has a Java Runtime Environment and was tested on most modern operating systems. Paxtools is open source and is available under the Lesser GNU public license (LGPL), which allows users to freely use the code in their software systems with a requirement for attribution. Source code for the current release (4.2.0) can be found in Software S1. A detailed manual for obtaining and using Paxtools can be found in Protocol S1. The latest sources and release bundles can be obtained from biopax.org/paxtools.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hu P, Bader G, Wigle DA, Emili A (2007) Computational prediction of cancer-gene function. Nature reviews Cancer 7: 23–34. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

