Structural analysis of the wheat genes encoding NADH-dependent glutamine-2-oxoglutarate amidotransferases and correlation with grain protein content
- PMID: 24069228
- PMCID: PMC3775782
- DOI: 10.1371/journal.pone.0073751
Structural analysis of the wheat genes encoding NADH-dependent glutamine-2-oxoglutarate amidotransferases and correlation with grain protein content
Abstract
Background: Nitrogen uptake and the efficient absorption and metabolism of nitrogen are essential elements in attempts to breed improved cereal cultivars for grain or silage production. One of the enzymes related to nitrogen metabolism is glutamine-2-oxoglutarate amidotransferase (GOGAT). Together with glutamine synthetase (GS), GOGAT maintains the flow of nitrogen from NH4 (+) into glutamine and glutamate, which are then used for several aminotransferase reactions during amino acid synthesis.
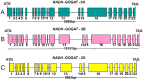
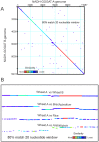
Results: The aim of the present work was to identify and analyse the structure of wheat NADH-GOGAT genomic sequences, and study the expression in two durum wheat cultivars characterized by low and high kernel protein content. The genomic sequences of the three homoeologous A, B and D NADH-GOGAT genes were obtained for hexaploid Triticum aestivum and the tetraploid A and B genes of Triticum turgidum ssp. durum. Analysis of the gene sequences indicates that all wheat NADH-GOGAT genes are composed of 22 exons and 21 introns. The three hexaploid wheat homoeologous genes have high conservation of sequence except intron 13 which shows differences in both length and sequence. A comparative analysis of sequences among di- and mono-cotyledonous plants shows both regions of high conservation and of divergence. qRT-PCR performed with the two durum wheat cvs Svevo and Ciccio (characterized by high and low protein content, respectively) indicates different expression levels of the two NADH-GOGAT-3A and NADH-GOGAT-3B genes.
Conclusion: The three hexaploid wheat homoeologous NADH-GOGAT gene sequences are highly conserved - consistent with the key metabolic role of this gene. However, the dicot and monocot amino acid sequences show distinctive patterns, particularly in the transit peptide, the exon 16-17 junction, and the C-terminus. The lack of conservation in the transit peptide may indicate subcellular differences between the two plant divisions - while the sequence conservation within enzyme functional domains remains high. Higher expression levels of NADH-GOGAT are associated with higher grain protein content in two durum wheats.
Conflict of interest statement
Figures





References
-
- Krapp A, Saliba-Colombani V, Daniel-Vedele F (2005) Analysis of C and N metabolisms and of C/N interactions using quantitative genetics. Photosyn Res 83: 251–263 doi:10.1007/s11120-004-3196-7 - DOI - PubMed
-
- Laperche A, Brancourt-Hulmel M, Heumez E, Gardet O, Le Gouis J (2006) Estimation of genetic parameters of a DH wheat population grown at different N stress levels characterized by probe genotypes. Theor Appl Genet 112: 797–807 doi:10.1007/s00122-005-0176-z - DOI - PubMed
-
- Hirel B, Le Gouis J, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58: 2369–2387 doi:10.1093/jxb/erm097 - DOI - PubMed
-
- Cren M, Hirel B (1999) Glutamine Synthetase in Higher Plants Regulation of Gene and Protein Expression from the Organ to the Cell. Plant Cell Physiol 40: 1187–1193.
-
- Ireland RJ, Lea PJ (1999) The enzymes of glutamine, glutamate, asparagines and aspartate metabolisms. Plant amino acids: Biochemistry and biotechnology. New York: Marcel Dekker. 49–109.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

