Genetic characterization of atypical Citrobacter freundii
- PMID: 24069274
- PMCID: PMC3771896
- DOI: 10.1371/journal.pone.0074120
Genetic characterization of atypical Citrobacter freundii
Abstract
The ability of a bacterial population to survive in different niches, as well as in stressful and rapidly changing environmental conditions, depends greatly on its genetic content. To survive such fluctuating conditions, bacteria have evolved different mechanisms to modulate phenotypic variations and related strategies to produce high levels of genetic diversity. Laboratories working in microbiological diagnosis have shown that Citrobacter freundii is very versatile in its colony morphology, as well as in its biochemical, antigenic and pathogenic behaviours. This phenotypic versatility has made C. freundii difficult to identify and it is frequently confused with both Salmonella enterica and Escherichia coli. In order to determine the genomic events and to explain the mechanisms involved in this plasticity, six C. freundii isolates were selected from a phenotypic variation study. An I-CeuI genomic cleavage map was created and eight housekeeping genes, including 16S rRNA, were sequenced. In general, the results showed a range of both phenotypes and genotypes among the isolates with some revealing a greater similarity to C. freundii and some to S. enterica, while others were identified as phenotypic and genotypic intermediary states between the two species. The occurrence of these events in natural populations may have important implications for genomic diversification in bacterial evolution, especially when considering bacterial species boundaries. In addition, such events may have a profound impact on medical science in terms of treatment, course and outcomes of infectious diseases, evading the immune response, and understanding host-pathogen interactions.
Conflict of interest statement
Figures


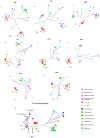

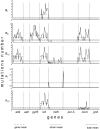
References
-
- Retchless AC, Lawrence JG (2007) Temporal fragmentation of speciation in bacteria. Science 317: 1093–1096. - PubMed
-
- Hallet B (2001) Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr Opin Microbiol 4: 570–581. - PubMed
-
- Veening JW, Smits WK, Kuipers OP (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62: 193–210. - PubMed
-
- Smits WK, Kuipers OP, Veening JW (2006) Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4: 259–271. - PubMed
-
- Dobrindt U, Hacker J (2001) Whole genome plasticity in pathogenic bacteria. Curr Opin Microbiol 4: 550–557. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

