Comparative genome analysis of Enterobacter cloacae
- PMID: 24069314
- PMCID: PMC3771936
- DOI: 10.1371/journal.pone.0074487
Comparative genome analysis of Enterobacter cloacae
Abstract
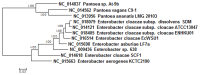
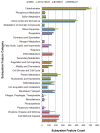
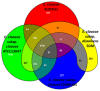
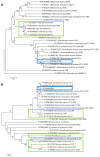
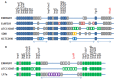
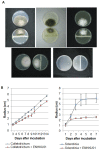
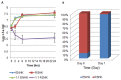
The Enterobacter cloacae species includes an extremely diverse group of bacteria that are associated with plants, soil and humans. Publication of the complete genome sequence of the plant growth-promoting endophytic E. cloacae subsp. cloacae ENHKU01 provided an opportunity to perform the first comparative genome analysis between strains of this dynamic species. Examination of the pan-genome of E. cloacae showed that the conserved core genome retains the general physiological and survival genes of the species, while genomic factors in plasmids and variable regions determine the virulence of the human pathogenic E. cloacae strain; additionally, the diversity of fimbriae contributes to variation in colonization and host determination of different E. cloacae strains. Comparative genome analysis further illustrated that E. cloacae strains possess multiple mechanisms for antagonistic action against other microorganisms, which involve the production of siderophores and various antimicrobial compounds, such as bacteriocins, chitinases and antibiotic resistance proteins. The presence of Type VI secretion systems is expected to provide further fitness advantages for E. cloacae in microbial competition, thus allowing it to survive in different environments. Competition assays were performed to support our observations in genomic analysis, where E. cloacae subsp. cloacae ENHKU01 demonstrated antagonistic activities against a wide range of plant pathogenic fungal and bacterial species.
Conflict of interest statement
Figures


Similar articles
-
Genome-wide Analysis of Four Enterobacter cloacae complex type strains: Insights into Virulence and Niche Adaptation.Sci Rep. 2020 May 18;10(1):8150. doi: 10.1038/s41598-020-65001-4. Sci Rep. 2020. PMID: 32424332 Free PMC article.
-
Complete genome sequence of the endophytic Enterobacter cloacae subsp. cloacae strain ENHKU01.J Bacteriol. 2012 Nov;194(21):5965. doi: 10.1128/JB.01394-12. J Bacteriol. 2012. PMID: 23045485 Free PMC article.
-
The whole-genome sequence analysis of Enterobacter cloacae strain Ghats1: insights into endophytic lifestyle-associated genomic adaptations.Arch Microbiol. 2020 Aug;202(6):1571-1579. doi: 10.1007/s00203-020-01848-5. Epub 2020 Mar 12. Arch Microbiol. 2020. PMID: 32166358
-
Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047.J Bacteriol. 2010 May;192(9):2463-4. doi: 10.1128/JB.00067-10. Epub 2010 Mar 5. J Bacteriol. 2010. PMID: 20207761 Free PMC article.
-
Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance.Future Microbiol. 2012 Jul;7(7):887-902. doi: 10.2217/fmb.12.61. Future Microbiol. 2012. PMID: 22827309 Review.
Cited by
-
Antihypertensive Therapy by ACEI/ARB Is Associated With Intestinal Flora Alterations and Metabolomic Profiles in Hypertensive Patients.Front Cell Dev Biol. 2022 Mar 23;10:861829. doi: 10.3389/fcell.2022.861829. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35399511 Free PMC article.
-
Enterobacter cloacae, an Emerging Plant-Pathogenic Bacterium Affecting Chili Pepper Seedlings.Plant Pathol J. 2018 Feb;34(1):1-10. doi: 10.5423/PPJ.OA.06.2017.0128. Epub 2018 Feb 1. Plant Pathol J. 2018. PMID: 29422783 Free PMC article.
-
Antibiotic Resistance Profile, Outer Membrane Proteins, Virulence Factors and Genome Sequence Analysis Reveal Clinical Isolates of Enterobacter Are Potential Pathogens Compared to Environmental Isolates.Front Cell Infect Microbiol. 2020 Feb 21;10:54. doi: 10.3389/fcimb.2020.00054. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32154188 Free PMC article.
-
Prevalence of Aminoglycoside Resistance Genes and Molecular Characterization of a Novel Gene, aac(3)-IIg, among Clinical Isolates of the Enterobacter cloacae Complex from a Chinese Teaching Hospital.Antimicrob Agents Chemother. 2020 Aug 20;64(9):e00852-20. doi: 10.1128/AAC.00852-20. Print 2020 Aug 20. Antimicrob Agents Chemother. 2020. PMID: 32571822 Free PMC article.
-
Multidrug-resistant phenotype and isolation of a novel SHV- beta-Lactamase variant in a clinical isolate of Enterobacter cloacae.J Biomed Sci. 2015 Apr 11;22(1):27. doi: 10.1186/s12929-015-0131-5. J Biomed Sci. 2015. PMID: 25888770 Free PMC article.
References
-
- Deangelis KM, D’Haeseleer P, Chivian D, Fortney JL, Khudyakov J et al. (2011) Complete genome sequence of "Enterobacter lignolyticus" SCF1. Standards Genomics Sciences 5: 69-85. doi:10.4056/sigs.2104875. - DOI - PMC - PubMed
-
- Zhu B, Lou MM, Xie GL, Wang GF, Zhou Q et al. (2011) Enterobacter mori sp. nov., associated with bacterial wilt on Morus alba L. Int J Syst Evol Microbiol 61: 2769-2774. doi:10.1099/ijs.0.028613-0. - DOI - PubMed
-
- Schroeder BK, Waters TD, du Toit LJ (2010) Evaluation of Onion Cultivars for Resistance to Enterobacter cloacae in Storage. Plant Dis 94: 236-243. doi:10.1094/PDIS-94-2-0236. - DOI - PubMed
-
- Liu S, Hu X, Lohrke SM, Baker CJ, Buyer JS et al. (2007) Role of sdhA and pfkA and catabolism of reduced carbon during colonization of cucumber roots by Enterobacter cloacae . Microbiology 153: 3196-3209. doi:10.1099/mic.0.2006/005538-0. - DOI - PubMed
-
- Hinton DM, Bacon CW (1995) Enterobacter cloacae is an endophytic symbiont of corn. Mycopathologia 129: 117-125. doi:10.1007/BF01103471. PubMed: 7659140. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

