Role for Krüppel-like transcription factor 11 in mesenchymal cell function and fibrosis
- PMID: 24069400
- PMCID: PMC3775729
- DOI: 10.1371/journal.pone.0075311
Role for Krüppel-like transcription factor 11 in mesenchymal cell function and fibrosis
Abstract
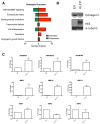
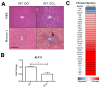
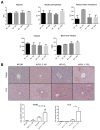
Krüppel-like factor 11 (KLF11) and the highly homologous KLF10 proteins are transcription factors originating from duplication of the Drosophila melanogaster ancestor cabut. The function of these proteins in epithelial cells has been previously characterized. In the current study, we report a functional role for KLF11 in mesenchymal cells and in mesenchymal cell dysfunction, namely, fibrosis, and subsequently perform a detailed cellular, molecular, and in vivo characterization of this phenomenon. We find that, in cultured mesenchymal cells, enhanced expression of KLF11 results in activated extracellular matrix pathways, including collagen gene silencing and matrix metalloproteinases activation without changes in tissue inhibitors of metalloproteinases. Combined, reporter and chromatin immunoprecipitation assays demonstrate that KLF11 interacts directly with the collagen 1a2 (COL1A2) promoter in mesenchymal cells to repress its activity. Mechanistically, KLF11 regulates collagen gene expression through the heterochromatin protein 1 gene-silencing pathway as mutants defective for coupling to this epigenetic modifier lose the ability to repress COL1A2. Expression studies reveal decreased levels of KLF11 during liver fibrogenesis after chemically induced injury in vivo. Congruently, KLF11(-/-) mice, which should be deficient in the hypothesized anti-fibrogenic brake imposed by this transcription factor, display an enhanced response to liver injury with increased collagen fibril deposition. Thus, KLFs expands the repertoire of transcription factors involved in the regulation of extracellular matrix proteins in mesenchymal cells and define a novel pathway that modulates the fibrogenic response during liver injury.
Conflict of interest statement
Figures



References
-
- Botella LM, Sánchez-Elsner T, Sanz-Rodriguez F, Kojima S, Shimada J et al. (2002) Transcriptional activation of endoglin and transforming growth factor-beta signaling components by cooperative interaction between Sp1 and KLF6: their potential role in the response to vascular injury. Blood 100: 4001-4010. PubMed: 12433697. - PubMed
-
- Botella LM, Sanz-Rodriguez F, Komi Y, Fernandez LA, Varela E et al. (2009) TGF-beta regulates the expression of transcription factor KLF6 and its splice variants and promotes co-operative transactivation of common target genes through a Smad3-Sp1-KLF6 interaction. Biochem J 419: 485-495. PubMed: 19076057. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

