A hybrid NRPS-PKS gene cluster related to the bleomycin family of antitumor antibiotics in Alteromonas macleodii strains
- PMID: 24069455
- PMCID: PMC3777966
- DOI: 10.1371/journal.pone.0076021
A hybrid NRPS-PKS gene cluster related to the bleomycin family of antitumor antibiotics in Alteromonas macleodii strains
Abstract
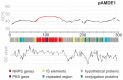
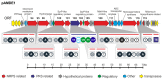
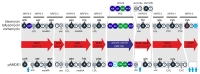
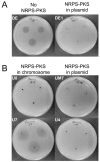
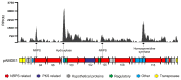
Although numerous marine bacteria are known to produce antibiotics via hybrid NRPS-PKS gene clusters, none have been previously described in an Alteromonas species. In this study, we describe in detail a novel hybrid NRPS-PKS cluster identified in the plasmid of the Alteromonasmacleodii strain AltDE1 and analyze its relatedness to other similar gene clusters in a sequence-based characterization. This is a mobile cluster, flanked by transposase-like genes, that has even been found inserted into the chromosome of some Alteromonasmacleodii strains. The cluster contains separate genes for NRPS and PKS activity. The sole PKS gene appears to carry a novel acyltransferase domain, quite divergent from those currently characterized. The predicted specificities of the adenylation domains of the NRPS genes suggest that the final compound has a backbone very similar to bleomycin related compounds. However, the lack of genes involved in sugar biosynthesis indicates that the final product is not a glycopeptide. Even in the absence of these genes, the presence of the cluster appears to confer complete or partial resistance to phleomycin, which may be attributed to a bleomycin-resistance-like protein identified within the cluster. This also suggests that the compound still shares significant structural similarity to bleomycin. Moreover, transcriptomic evidence indicates that the NRPS-PKS cluster is expressed. Such sequence-based approaches will be crucial to fully explore and analyze the diversity and potential of secondary metabolite production, especially from increasingly important sources like marine microbes.
Conflict of interest statement
Figures

References
-
- Blunt JW, Copp BR, Hu WP, Munro MH, Northcote PT et al. (2009) Marine natural products. Nat Prod Rep 26: 170-244. doi:10.1039/b805113p. PubMed: 19177222. - DOI - PubMed
-
- Li JWH, Vederas JC (2009) Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 325: 161-165. doi:10.1126/science.1168243. PubMed: 19589993. - DOI - PubMed
-
- Gulder TAM, Moore BS (2009) Chasing the treasures of the sea - bacterial marine natural products. Curr Opin Microbiol 12: 252-260. doi:10.1016/j.mib.2009.05.002. PubMed: 19481972. - DOI - PMC - PubMed
-
- Du L, Sánchez C, Shen B (2001) Hybrid Peptide–Polyketide Natural Products: Biosynthesis and Prospects toward Engineering Novel Molecules. Metab Eng 3: 78-95. doi:10.1006/mben.2000.0171. PubMed: 11162234. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

