Identification of TET1 Partners That Control Its DNA-Demethylating Function
- PMID: 24069510
- PMCID: PMC3782005
- DOI: 10.1177/1947601913489020
Identification of TET1 Partners That Control Its DNA-Demethylating Function
Abstract
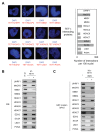
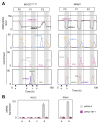
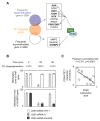
Several recent reports have identified TET1 as the main enzyme modulating DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. However, little is known about the protein network that controls TET1 activity. By using a new proximity ligation in situ assay, we identified MeCP2, HDAC1/6/7, EZH2, mSin3A, PCNA, and LSD1 as TET1-interacting proteins. We also discerned that TET1/PCNA acts as a demethylator of the cyclical methylation/demethylation process, the perturbation of which promotes the aberrant methylation hallmarks frequently observed in cancer cells.
Keywords: DNA methylation; TET1; demethylation; glioma.
Conflict of interest statement
Figures

Similar articles
-
Alterations of regulatory factors and DNA methylation pattern in thyroid cancer.Cancer Biomark. 2020;28(2):255-268. doi: 10.3233/CBM-190871. Cancer Biomark. 2020. PMID: 32390600
-
TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation.Neuron. 2013 Sep 18;79(6):1086-93. doi: 10.1016/j.neuron.2013.08.032. Neuron. 2013. PMID: 24050399 Free PMC article.
-
TET1 exerts its tumor suppressor function by interacting with p53-EZH2 pathway in gastric cancer.J Biomed Nanotechnol. 2014 Jul;10(7):1217-30. doi: 10.1166/jbn.2014.1861. J Biomed Nanotechnol. 2014. PMID: 24804542
-
Aberrant TET1 Methylation Closely Associated with CpG Island Methylator Phenotype in Colorectal Cancer.Cancer Prev Res (Phila). 2015 Aug;8(8):702-11. doi: 10.1158/1940-6207.CAPR-14-0306. Epub 2015 Jun 10. Cancer Prev Res (Phila). 2015. PMID: 26063725
-
Multiple Functions of Ten-eleven Translocation 1 during Tumorigenesis.Chin Med J (Engl). 2016 Jul 20;129(14):1744-51. doi: 10.4103/0366-6999.185873. Chin Med J (Engl). 2016. PMID: 27411465 Free PMC article. Review.
Cited by
-
Hepatitis B virus X protein induces EpCAM expression via active DNA demethylation directed by RelA in complex with EZH2 and TET2.Oncogene. 2016 Feb 11;35(6):715-26. doi: 10.1038/onc.2015.122. Epub 2015 Apr 20. Oncogene. 2016. PMID: 25893293 Free PMC article.
-
CTCF as a regulator of alternative splicing: new tricks for an old player.Nucleic Acids Res. 2021 Aug 20;49(14):7825-7838. doi: 10.1093/nar/gkab520. Nucleic Acids Res. 2021. PMID: 34181707 Free PMC article. Review.
-
Modification of Tet1 and histone methylation dynamics in dairy goat male germline stem cells.Cell Prolif. 2016 Apr;49(2):163-72. doi: 10.1111/cpr.12245. Epub 2016 Mar 14. Cell Prolif. 2016. PMID: 26988797 Free PMC article.
-
Rett syndrome - biological pathways leading from MECP2 to disorder phenotypes.Orphanet J Rare Dis. 2016 Nov 25;11(1):158. doi: 10.1186/s13023-016-0545-5. Orphanet J Rare Dis. 2016. PMID: 27884167 Free PMC article. Review.
-
MeCP2 regulates Tet1-catalyzed demethylation, CTCF binding, and learning-dependent alternative splicing of the BDNF gene in Turtle.Elife. 2017 Jun 8;6:e25384. doi: 10.7554/eLife.25384. Elife. 2017. PMID: 28594324 Free PMC article.
References
-
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi Y. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390-3 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
