α-helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes
- PMID: 24071712
- PMCID: PMC3784961
- DOI: 10.1038/srep02781
α-helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes
Abstract
The human islet amyloid polypeptide (hIAPP) is the primary component in the toxic islet amyloid deposits in type-2 diabetes. hIAPP self-assembles to aggregates that permeabilize membranes and constitutes amyloid plaques. Uncovering the mechanisms of amyloid self-assembly is the key to understanding amyloid toxicity and treatment. Although structurally similar, hIAPP's rat counterpart, the rat islet amyloid polypeptide (rIAPP), is non-toxic. It has been a puzzle why these peptides behave so differently. We combined multiscale modelling and theory to explain the drastically different dynamics of hIAPP and rIAPP: The differences stem from electrostatic dipolar interactions. hIAPP forms pentameric aggregates with the hydrophobic residues facing the membrane core and stabilizing water-conducting pores. We give predictions for pore sizes, the number of hIAPP peptides, and aggregate morphology. We show the importance of curvature-induced stress at the early stages of hIAPP assembly and the α-helical structures over β-sheets. This agrees with recent fluorescence spectroscopy experiments.
Figures
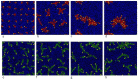
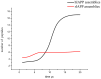

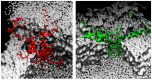

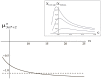
 . From the top to the bottom:
. From the top to the bottom:  = 1, 2, 3.
= 1, 2, 3.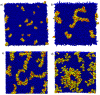

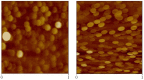
Similar articles
-
Structural and energetic insight into the cross-seeding amyloid assemblies of human IAPP and rat IAPP.J Phys Chem B. 2014 Jun 26;118(25):7026-36. doi: 10.1021/jp5022246. Epub 2014 Jun 12. J Phys Chem B. 2014. PMID: 24892388
-
Membrane Interactions of hIAPP Monomer and Oligomer with Lipid Membranes by Molecular Dynamics Simulations.ACS Chem Neurosci. 2017 Aug 16;8(8):1789-1800. doi: 10.1021/acschemneuro.7b00160. Epub 2017 Jun 13. ACS Chem Neurosci. 2017. PMID: 28585804
-
Islet amyloid polypeptide: aggregation and fibrillogenesis in vitro and its inhibition.Subcell Biochem. 2012;65:185-209. doi: 10.1007/978-94-007-5416-4_8. Subcell Biochem. 2012. PMID: 23225004 Review.
-
Comparative molecular dynamics study of human islet amyloid polypeptide (IAPP) and rat IAPP oligomers.Biochemistry. 2013 Feb 12;52(6):1089-100. doi: 10.1021/bi301525e. Epub 2013 Jan 29. Biochemistry. 2013. PMID: 23331123
-
Human islet amyloid polypeptide (hIAPP) - a curse in type II diabetes mellitus: insights from structure and toxicity studies.Biol Chem. 2020 Sep 4;402(2):133-153. doi: 10.1515/hsz-2020-0174. Print 2021 Jan 27. Biol Chem. 2020. PMID: 33544470 Review.
Cited by
-
Aggregation of Chameleon Peptides: Implications of α-Helicity in Fibril Formation.J Phys Chem B. 2016 Jul 7;120(26):5874-83. doi: 10.1021/acs.jpcb.6b00830. Epub 2016 Apr 1. J Phys Chem B. 2016. PMID: 27001160 Free PMC article.
-
Growth kinetics of amyloid-like fibrils: An integrated atomistic simulation and continuum theory approach.PNAS Nexus. 2024 Feb 1;3(2):pgae045. doi: 10.1093/pnasnexus/pgae045. eCollection 2024 Feb. PNAS Nexus. 2024. PMID: 38725528 Free PMC article.
-
Characterisation of the Structure and Oligomerisation of Islet Amyloid Polypeptides (IAPP): A Review of Molecular Dynamics Simulation Studies.Molecules. 2018 Aug 25;23(9):2142. doi: 10.3390/molecules23092142. Molecules. 2018. PMID: 30149632 Free PMC article. Review.
-
Dual Effects of Presynaptic Membrane Mimetics on α-Synuclein Amyloid Aggregation.Front Cell Dev Biol. 2022 Jun 7;10:707417. doi: 10.3389/fcell.2022.707417. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35747692 Free PMC article.
-
Mechanism of Inhibition of Human Islet Amyloid Polypeptide-Induced Membrane Damage by a Small Organic Fluorogen.Sci Rep. 2016 Feb 18;6:21614. doi: 10.1038/srep21614. Sci Rep. 2016. PMID: 26887358 Free PMC article.
References
-
- Merlini G. & Bellotti V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 349, 583–596 (2003). - PubMed
-
- Hardy J. & Selkoe D. J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002). - PubMed
-
- Dickson D. W. The pathogenesis of senile plaques. J. Neuropathol. Exp. Neurol. 56, 321–339 (1997). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

