Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas
- PMID: 24071850
- PMCID: PMC3950337
- DOI: 10.1038/ng.2761
Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas
Abstract
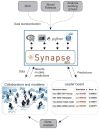
The Cancer Genome Atlas Pan-Cancer Analysis Working Group collaborated on the Synapse software platform to share and evolve data, results and methodologies while performing integrative analysis of molecular profiling data from 12 tumor types. The group's work serves as a pilot case study that provides (i) a template for future large collaborative studies; (ii) a system to support collaborative projects; and (iii) a public resource of highly curated data, results and automated systems for the evaluation of community-developed models.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

