Presence-absence variation in A. thaliana is primarily associated with genomic signatures consistent with relaxed selective constraints
- PMID: 24072814
- PMCID: PMC3879440
- DOI: 10.1093/molbev/mst166
Presence-absence variation in A. thaliana is primarily associated with genomic signatures consistent with relaxed selective constraints
Abstract
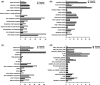
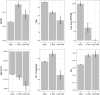
The sequencing of multiple genomes of the same plant species has revealed polymorphic gene and exon loss. Genes associated with disease resistance are overrepresented among those showing structural variations, suggesting an adaptive role for gene and exon presence-absence variation (PAV). To shed light on the possible functional relevance of polymorphic coding region loss and the mechanisms driving this process, we characterized genes that have lost entire exons or their whole coding regions in 17 fully sequenced Arabidopsis thaliana accessions. We found that although a significant enrichment in genes associated with certain functional categories is observed, PAV events are largely restricted to genes with signatures of reduced essentiality: PAV genes tend to be newer additions to the genome, tissue specific, and lowly expressed. In addition, PAV genes are located in regions of lower gene density and higher transposable element density. Partial coding region PAV events were associated with only a marginal reduction in gene expression level in the affected accession and occurred in genes with higher levels of alternative splicing in the Col-0 accession. Together, these results suggest that although adaptive scenarios cannot be ruled out, PAV events can be explained without invoking them.
Keywords: Arabidopsis; adaptive evolution; exon deletion; presence–absence variation; transposable elements; whole genome evolution.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST—database for expressed sequence tags. Nat Genet. 1993;4:332–333. - PubMed
-
- Brenner S, Johnson M, Bridgham J, et al. (24 co-authors) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. - PubMed
-
- Cao J, Schneeberger K, Ossowski S, et al. (17 co-authors) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–963. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

