Approaches for assessing and discovering protein interactions in cancer
- PMID: 24072816
- PMCID: PMC3834224
- DOI: 10.1158/1541-7786.MCR-13-0454
Approaches for assessing and discovering protein interactions in cancer
Abstract
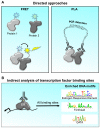
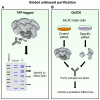
Significant insight into the function of proteins can be delineated by discovering and characterizing interacting proteins. There are numerous methods for the discovery of unknown associated protein networks, with purification of the bait (the protein of interest) followed by mass spectrometry as a common theme. In recent years, advances have permitted the purification of endogenous proteins and methods for scaling down starting material. As such, approaches for rapid, unbiased identification of protein interactomes are becoming a standard tool in the researchers toolbox, rather than a technique that is only available to specialists. This review will highlight some of the recent technical advances in proteomic-based discovery approaches, the pros and cons of various methods and some of the key findings in cancer-related systems.
©2013 AACR.
Figures

References
-
- L’Ecuyer TJ, Fulton AB. Specific and quantitative immunoprecipitation of tropomyosin and other cytoskeletal proteins by magnetic separation. BioTechniques. 1993;14:436–41. - PubMed
-
- Day RN. Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy. Molecular endocrinology. 1998;12:1410–9. - PubMed
-
- Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

