Differential host response, rather than early viral replication efficiency, correlates with pathogenicity caused by influenza viruses
- PMID: 24073225
- PMCID: PMC3779241
- DOI: 10.1371/journal.pone.0074863
Differential host response, rather than early viral replication efficiency, correlates with pathogenicity caused by influenza viruses
Abstract
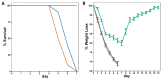
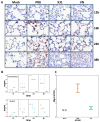
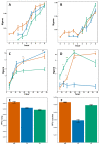
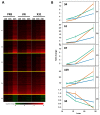
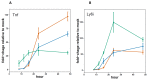
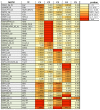
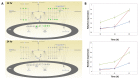
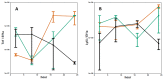
Influenza viruses exhibit large, strain-dependent differences in pathogenicity in mammalian hosts. Although the characteristics of severe disease, including uncontrolled viral replication, infection of the lower airway, and highly inflammatory cytokine responses have been extensively documented, the specific virulence mechanisms that distinguish highly pathogenic strains remain elusive. In this study, we focused on the early events in influenza infection, measuring the growth rate of three strains of varying pathogenicity in the mouse airway epithelium and simultaneously examining the global host transcriptional response over the first 24 hours. Although all strains replicated equally rapidly over the first viral life-cycle, their growth rates in both lung and tracheal tissue strongly diverged at later times, resulting in nearly 10-fold differences in viral load by 24 hours following infection. We identified separate networks of genes in both the lung and tracheal tissues whose rapid up-regulation at early time points by specific strains correlated with a reduced viral replication rate of those strains. The set of early-induced genes in the lung that led to viral growth restriction is enriched for both NF-κB binding site motifs and members of the TREM1 and IL-17 signaling pathways, suggesting that rapid, NF-κB -mediated activation of these pathways may contribute to control of viral replication. Because influenza infection extending into the lung generally results in severe disease, early activation of these pathways may be one factor distinguishing high- and low-pathogenicity strains.
Conflict of interest statement
Figures

References
-
- WHO (2009) Influenza (Seasonal) Fact Sheet No 211. World Health Organization.
-
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM et al. (2007) The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25: 5086-5096. doi:10.1016/j.vaccine.2007.03.046. PubMed: 17544181. - DOI - PubMed
-
- Meltzer MI, Cox NJ, Fukuda K (1999) The economic impact of pandemic influenza in the United States: priorities for intervention. Emerg Infect Dis 5: 659-671. doi:10.3201/eid0505.990507. PubMed: 10511522. - DOI - PMC - PubMed
-
- Johnson NP, Mueller J (2002) Updating the accounts: global mortality of the 1918-1920 "Spanish" influenza pandemic. Bull Hist Med 76: 105-115. doi:10.1353/bhm.2002.0022. PubMed: 11875246. - DOI - PubMed
-
- Faruqui F, Mukundan D (2010) 2009 pandemic influenza: a review. Curr Opin Pediatr 22: 530-535. doi:10.1097/MOP.0b013e32833bb81a. PubMed: 20601883. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

