Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress
- PMID: 24074255
- PMCID: PMC3849779
- DOI: 10.1186/1471-2164-14-662
Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress
Abstract
Background: Chrysanthemum is one of the most important ornamental crops in the world and drought stress seriously limits its production and distribution. In order to generate a functional genomics resource and obtain a deeper understanding of the molecular mechanisms regarding chrysanthemum responses to dehydration stress, we performed large-scale transcriptome sequencing of chrysanthemum plants under dehydration stress using the Illumina sequencing technology.
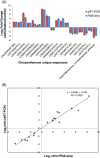
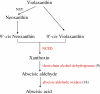
Results: Two cDNA libraries constructed from mRNAs of control and dehydration-treated seedlings were sequenced by Illumina technology. A total of more than 100 million reads were generated and de novo assembled into 98,180 unique transcripts which were further extensively annotated by comparing their sequencing to different protein databases. Biochemical pathways were predicted from these transcript sequences. Furthermore, we performed gene expression profiling analysis upon dehydration treatment in chrysanthemum and identified 8,558 dehydration-responsive unique transcripts, including 307 transcription factors and 229 protein kinases and many well-known stress responsive genes. Gene ontology (GO) term enrichment and biochemical pathway analyses showed that dehydration stress caused changes in hormone response, secondary and amino acid metabolism, and light and photoperiod response. These findings suggest that drought tolerance of chrysanthemum plants may be related to the regulation of hormone biosynthesis and signaling, reduction of oxidative damage, stabilization of cell proteins and structures, and maintenance of energy and carbon supply.
Conclusions: Our transcriptome sequences can provide a valuable resource for chrysanthemum breeding and research and novel insights into chrysanthemum responses to dehydration stress and offer candidate genes or markers that can be used to guide future studies attempting to breed drought tolerant chrysanthemum cultivars.
Figures






References
-
- Teixeira Da Silva JA. Chrysanthemum: advances in tissue culture, cryopreservation, postharvest technology, genetics and transgenic biotechnology. Biotechnol Adv. 2003;21:715–766. - PubMed
-
- Anderson NO. In: In Flower Breeding and Genetics. Anderson NO, editor. Netherlands: Springer; 2006. Chrysanthemum; pp. 389–437.
-
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

