Mechanisms of HIV-1 subtype C resistance to GRFT, CV-N and SVN
- PMID: 24074568
- PMCID: PMC3787538
- DOI: 10.1016/j.virol.2013.07.019
Mechanisms of HIV-1 subtype C resistance to GRFT, CV-N and SVN
Abstract
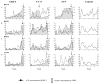
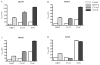
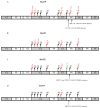
We examined the ability of HIV-1 subtype C to develop resistance to the inhibitory lectins, griffithsin (GRFT), cyanovirin-N (CV-N) and scytovirin (SVN), which bind multiple mannose-rich glycans on gp120. Four primary HIV-1 strains cultured under escalating concentrations of these lectins became increasingly resistant tolerating 2 to 12 times their 50% inhibitory concentrations. Sequence analysis of gp120 showed that most had deletions of 1 to 5 mannose-rich glycans. Glycosylation sites at positions 230, 234, 241, 289 located in the C2 region and 339, 392 and 448 in the C3-C4 region were affected. Furthermore, deletions and insertions of up to 5 amino acids in the V4 region were observed in 3 of the 4 isolates. These data suggest that loss of glycosylation sites on gp120 as well as rearrangement of glycans in V4 are mechanisms involved in HIV-1 subtype C escape from GRFT, CV-N and SVN.
Keywords: Cyanovirin-N; Entry inhibitor; Glycans; Griffithsin; HIV subtype C; Microbicide; Resistance; Scytovirin; Single genome amplification.
© 2013 Elsevier Inc. All rights reserved.
Figures


References
-
- Balzarini J. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infect Dis. 2005;5:726–731. - PubMed
-
- Balzarini J, Van Laethem K, Hatse S, Froeyen M, Van Damme E, Bolmstedt A, Peumans W, De Clercq E, Schols D. Marked depletion of glycosylation sites in HIV-1 gp120 under selection pressure by the mannose-specific plant lectins of Hippeastrum hybrid and Galanthus nivalis. Mol Pharmacol. 2005;67:1556–1565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

