Mapping human epigenomes
- PMID: 24074860
- PMCID: PMC3838898
- DOI: 10.1016/j.cell.2013.09.011
Mapping human epigenomes
Abstract
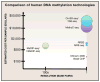
As the second dimension to the genome, the epigenome contains key information specific to every type of cells. Thousands of human epigenome maps have been produced in recent years thanks to rapid development of high throughput epigenome mapping technologies. In this review, we discuss the current epigenome mapping toolkit and utilities of epigenome maps. We focus particularly on mapping of DNA methylation, chromatin modification state, and chromatin structures, and emphasize the use of epigenome maps to delineate human gene regulatory sequences and developmental programs. We also provide a perspective on the progress of the epigenomics field and challenges ahead.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
References
-
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, Van Lohuizen M, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials