A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk
- PMID: 24074861
- PMCID: PMC3844554
- DOI: 10.1016/j.cell.2013.08.030
A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk
Abstract
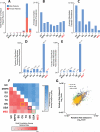
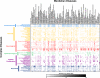
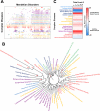
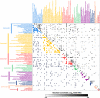
Although countless highly penetrant variants have been associated with Mendelian disorders, the genetic etiologies underlying complex diseases remain largely unresolved. By mining the medical records of over 110 million patients, we examine the extent to which Mendelian variation contributes to complex disease risk. We detect thousands of associations between Mendelian and complex diseases, revealing a nondegenerate, phenotypic code that links each complex disorder to a unique collection of Mendelian loci. Using genome-wide association results, we demonstrate that common variants associated with complex diseases are enriched in the genes indicated by this "Mendelian code." Finally, we detect hundreds of comorbidity associations among Mendelian disorders, and we use probabilistic genetic modeling to demonstrate that Mendelian variants likely contribute nonadditively to the risk for a subset of complex diseases. Overall, this study illustrates a complementary approach for mapping complex disease loci and provides unique predictions concerning the etiologies of specific diseases.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Comment in
-
Disease genetics: a Mendelian code for complex disease.Nat Rev Genet. 2013 Nov;14(11):746. doi: 10.1038/nrg3599. Nat Rev Genet. 2013. PMID: 24136496 No abstract available.
References
-
- Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nature reviews Genetics. 2002;3:779–789. - PubMed
-
- Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, Lewis RA, Beales PL, Dietz HC, Fisher S, Katsanis N. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. - PubMed
-
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;10:10008–10020.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

