Genetic variants regulating immune cell levels in health and disease
- PMID: 24074872
- PMCID: PMC5541764
- DOI: 10.1016/j.cell.2013.08.041
Genetic variants regulating immune cell levels in health and disease
Abstract
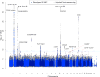
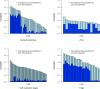
The complex network of specialized cells and molecules in the immune system has evolved to defend against pathogens, but inadvertent immune system attacks on "self" result in autoimmune disease. Both genetic regulation of immune cell levels and their relationships with autoimmunity are largely undetermined. Here, we report genetic contributions to quantitative levels of 95 cell types encompassing 272 immune traits, in a cohort of 1,629 individuals from four clustered Sardinian villages. We first estimated trait heritability, showing that it can be substantial, accounting for up to 87% of the variance (mean 41%). Next, by assessing ∼8.2 million variants that we identified and confirmed in an extended set of 2,870 individuals, 23 independent variants at 13 loci associated with at least one trait. Notably, variants at three loci (HLA, IL2RA, and SH2B3/ATXN2) overlap with known autoimmune disease associations. These results connect specific cellular phenotypes to specific genetic variants, helping to explicate their involvement in disease.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Auer PL, Johnsen JM, Johnson AD, Logsdon BA, Lange LA, Nalls MA, Zhang G, Franceschini N, Fox K, Lange EM, et al. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am J Hum Genet. 2012;91:794–808. - PMC - PubMed
-
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. - PubMed
-
- Bayés B, Pastor MC, Lauzurica R, Granada ML, Salinas I, Romero R. Do anti-CD25 monoclonal antibodies potentiate posttransplant diabetes mellitus? Transplant Proc. 2007;39:2248–2250. - PubMed
-
- Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, Giometto R, Höpner S, Centonze D, Bernardi G, Dell’Acqua ML, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. 2007;110:1225–1232. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

