Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly
- PMID: 24074955
- PMCID: PMC3929958
- DOI: 10.1016/j.molcel.2013.08.032
Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly
Abstract
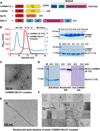
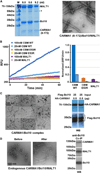
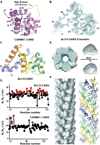
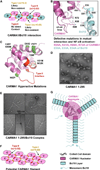
The CARMA1/Bcl10/MALT1 (CBM) signalosome mediates antigen receptor-induced NF-κB signaling to regulate multiple lymphocyte functions. While CARMA1 and Bcl10 contain caspase recruitment domains (CARDs), MALT1 is a paracaspase with structural similarity to caspases. Here we show that the reconstituted CBM signalosome is a helical filamentous assembly in which substoichiometric CARMA1 nucleates Bcl10 filaments. Bcl10 filament formation is a highly cooperative process whose threshold is sensitized by oligomerized CARMA1 upon receptor activation. In cells, both cotransfected CARMA1/Bcl10 complex and the endogenous CBM signalosome are filamentous morphologically. Combining crystallography, nuclear magnetic resonance, and electron microscopy, we reveal the structure of the Bcl10 CARD filament and the mode of interaction between CARMA1 and Bcl10. Structure-guided mutagenesis confirmed the observed interfaces in Bcl10 filament assembly and MALT1 activation in vitro and NF-κB activation in cells. These data support a paradigm of nucleation-induced signal transduction with threshold response due to cooperativity and signal amplification by polymerization.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


References
-
- Bertin J, Guo Y, Wang L, Srinivasula SM, Jacobson MD, Poyet JL, Merriam S, Du MQ, Dyer MJ, Robison KE, et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J. Biol. Chem. 2000;275:41082–41086. - PubMed
-
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat. Immunol. 2008;9:263–271. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

