The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit
- PMID: 24075009
- PMCID: PMC4001917
- DOI: 10.1016/j.cell.2013.08.062
The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit
Abstract
Tissue homeostasis in metazoans is regulated by transitions of cells between quiescence and proliferation. The hallmark of proliferating populations is progression through the cell cycle, which is driven by cyclin-dependent kinase (CDK) activity. Here, we introduce a live-cell sensor for CDK2 activity and unexpectedly found that proliferating cells bifurcate into two populations as they exit mitosis. Many cells immediately commit to the next cell cycle by building up CDK2 activity from an intermediate level, while other cells lack CDK2 activity and enter a transient state of quiescence. This bifurcation is directly controlled by the CDK inhibitor p21 and is regulated by mitogens during a restriction window at the end of the previous cell cycle. Thus, cells decide at the end of mitosis to either start the next cell cycle by immediately building up CDK2 activity or to enter a transient G0-like state by suppressing CDK2 activity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

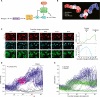
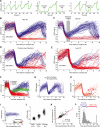
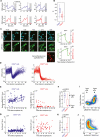
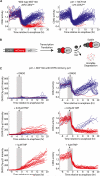

Comment in
-
Cell cycle: Making an early decision.Nat Rev Mol Cell Biol. 2013 Nov;14(11):691. doi: 10.1038/nrm3687. Epub 2013 Oct 16. Nat Rev Mol Cell Biol. 2013. PMID: 24129417 No abstract available.
References
-
- Bachman KE, Blair BG, Brenner K, Bardelli A, Arena S, Zhou S, Hicks J, De Marzo AM, Argani P, Park BH. p21(WAF1/CIP1) mediates the growth response to TGF-beta in human epithelial cells. Cancer Biol. Ther. 2004;3:221–225. - PubMed
-
- Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. - PubMed
-
- Chassot AA, Lossaint G, Turchi L, Meneguzzi G, Fisher D, Ponzio G, Dulic V. Confluence-induced cell cycle exit involves pre-mitotic CDK inhibition by p27(Kip1) and cyclin D1 downregulation. Cell Cycle. 2008;7:2038–2046. - PubMed
-
- Chen PL, Scully P, Shew JY, Wang JY, Lee WH. Phosphor-ylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

