Population genomics of rapid adaptation by soft selective sweeps
- PMID: 24075201
- PMCID: PMC3834262
- DOI: 10.1016/j.tree.2013.08.003
Population genomics of rapid adaptation by soft selective sweeps
Abstract
Organisms can often adapt surprisingly quickly to evolutionary challenges, such as the application of pesticides or antibiotics, suggesting an abundant supply of adaptive genetic variation. In these situations, adaptation should commonly produce 'soft' selective sweeps, where multiple adaptive alleles sweep through the population at the same time, either because the alleles were already present as standing genetic variation or arose independently by recurrent de novo mutations. Most well-known examples of rapid molecular adaptation indeed show signatures of such soft selective sweeps. Here, we review the current understanding of the mechanisms that produce soft sweeps and the approaches used for their identification in population genomic data. We argue that soft sweeps might be the dominant mode of adaptation in many species.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures



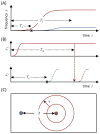
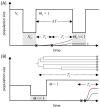
References
-
- Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002;3:11–21. - PubMed
-
- Fisher R. The genetical theory of natural selection. Clarendon Press; Oxford: 1930.
-
- Daborn P, et al. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol Genet Genomics. 2001;266:556–563. - PubMed
-
- Hoekstra HE, et al. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. - PubMed
-
- Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

