Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma
- PMID: 24075231
- PMCID: PMC3981552
- DOI: 10.1016/j.jaci.2013.07.036
Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma
Abstract
Background: The 3-gene signature of periostin, chloride channel accessory 1 (CLCA1), and Serpin β2 (SERPINB2) in airway epithelial brushings is used to classify asthma into TH2-high and TH2-low endotypes. Little is known about the utility of gene profiling in sputum as a molecular phenotyping method.
Objective: We sought to determine whether gene profiling in sputum cells can identify T(H)2-high and T(H)2-low subtypes of asthma.
Methods: In induced sputum cell pellets from 37 asthmatic patients and 15 healthy control subjects, PCR was used to profile gene expression of the epithelial cell signature of IL-13 activation (periostin, CLCA1, and SERPINB2), TH2 genes (IL4, IL5, and IL13), and other genes associated with airway TH2 inflammation.
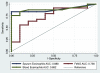
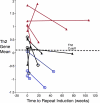
Results: Gene expression levels of CLCA1 and periostin, but not SerpinB2, were significantly higher than normal in sputum cells from asthmatic subjects. Expression levels of IL-4, IL-5, and IL-13 were also significantly increased in asthmatic patients and highly correlated within individual subjects. By combining the expression levels of IL-4, IL-5, and IL-13 in a single quantitative metric ("T(H)2 gene mean"), 26 (70%) of the 37 asthmatic patients had T(H)2-high asthma, which was characterized by more severe measures of asthma and increased blood and sputum eosinophilia. TH2 gene mean values tended to be stable when initial values were very high or very low but fluctuated above or below the T(H)2-high cutoff when initial values were intermediate.
Conclusion: IL-4, IL-5, and IL-13 transcripts are easily detected in sputum cells from asthmatic patients, and their expression levels can be used to classify asthma into T(H)2-high and T(H)2-low endotypes.
Keywords: Asthma; BME; CLCA1; CPA3; Carboxypeptidase A3; Chloride channel accessory 1; Feno; Fraction of exhaled nitric oxide; IL-13; IL-17; IL-4; IL-5; LABA; Long-acting β-agonist; RIN; RNA integrity number; Serpin β2; SerpinB2; T(H)2 cell; UCSF; University of California, San Francisco; cytokines; eosinophils; eotaxin; inflammation; mast cells; phenotypes; sputum; β-Mercaptoethanol.
Copyright © 2013 American Academy of Allergy, Asthma & Immunology. Published by Mosby, Inc. All rights reserved.
Figures





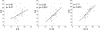
Comment in
-
Asthma phenotyping: TH2-high, TH2-low, and beyond.J Allergy Clin Immunol. 2014 Feb;133(2):395-6. doi: 10.1016/j.jaci.2013.10.008. Epub 2013 Nov 28. J Allergy Clin Immunol. 2014. PMID: 24290288 No abstract available.
References
-
- Robinson DS, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bentley AM, et al. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med. 1992;326:298–304. - PubMed
-
- Corren J, Busse W, Meltzer EO, Mansfield L, Bensch G, Fahrenholz J, et al. A randomized, controlled, phase 2 study of AMG 317, an IL-4Ralpha antagonist, in patients with asthma. Am J Respir Crit Care Med. 2010;181:788–96. - PubMed
-
- Kips JC, O'Connor BJ, Langley SJ, Woodcock A, Kerstjens HA, Postma DS, et al. Effect of SCH55700, a humanized anti-human interleukin-5 antibody, in severe persistent asthma: a pilot study. Am J Respir Crit Care Med. 2003;167:1655–9. - PubMed
-
- Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009;360:985–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

