Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells
- PMID: 24075860
- PMCID: PMC3837626
- DOI: 10.1016/j.chom.2013.09.002
Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells
Abstract
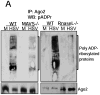
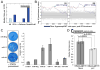
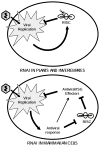
RNA interference (RNAi) is an established antiviral defense mechanism in plants and invertebrates. Whether RNAi serves a similar function in mammalian cells remains unresolved. We find that in some cell types, mammalian RNAi activity is reduced shortly after viral infection via poly-ADP-ribosylation of the RNA-induced silencing complex (RISC), a core component of RNAi. Well-established antiviral signaling pathways, including RIG-I/MAVS and RNaseL, contribute to inhibition of RISC. In the absence of virus infection, microRNAs repress interferon-stimulated genes (ISGs) associated with cell death and proliferation, thus maintaining homeostasis. Upon detection of intracellular pathogen-associated molecular patterns, RISC activity decreases, contributing to increased expression of ISGs. Our results suggest that, unlike in lower eukaryotes, mammalian RISC is not antiviral in some contexts, but rather RISC has been co-opted to negatively regulate toxic host antiviral effectors via microRNAs.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Barber GN. The interferons and cell death: guardians of the cell or accomplices of apoptosis? Semin Cancer Biol. 2000;10:103–111. - PubMed
-
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. - PubMed
-
- Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, Borden EC. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

