Dendritic glutamate receptor mRNAs show contingent local hotspot-dependent translational dynamics
- PMID: 24075992
- PMCID: PMC3901016
- DOI: 10.1016/j.celrep.2013.08.029
Dendritic glutamate receptor mRNAs show contingent local hotspot-dependent translational dynamics
Abstract
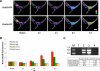
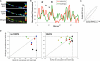
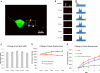
Protein synthesis in neuronal dendrites underlies long-term memory formation in the brain. Local translation of reporter mRNAs has demonstrated translation in dendrites at focal points called translational hotspots. Various reports have shown that hundreds to thousands of mRNAs are localized to dendrites, yet the dynamics of translation of multiple dendritic mRNAs has remained elusive. Here, we show that the protein translational activities of two dendritically localized mRNAs are spatiotemporally complex but constrained by the translational hotspots in which they are colocalized. Cotransfection of glutamate receptor 2 (GluR2) and GluR4 mRNAs (engineered to encode different fluorescent proteins) into rat hippocampal neurons demonstrates a heterogeneous distribution of translational hotspots for the two mRNAs along dendrites. Stimulation with s-3,5-dihydroxy-phenylglycine modifies the translational dynamics of both of these RNAs in a complex saturable manner. These results suggest that the translational hotspot is a primary structural regulator of the simultaneous yet differential translation of multiple mRNAs in the neuronal dendrite.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

