Circadian and circalunar clock interactions in a marine annelid
- PMID: 24075994
- PMCID: PMC3913041
- DOI: 10.1016/j.celrep.2013.08.031
Circadian and circalunar clock interactions in a marine annelid
Abstract
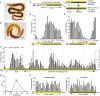
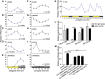
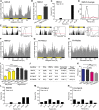
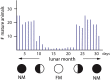
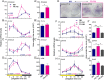
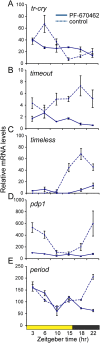
Life is controlled by multiple rhythms. Although the interaction of the daily (circadian) clock with environmental stimuli, such as light, is well documented, its relationship to endogenous clocks with other periods is little understood. We establish that the marine worm Platynereis dumerilii possesses endogenous circadian and circalunar (monthly) clocks and characterize their interactions. The RNAs of likely core circadian oscillator genes localize to a distinct nucleus of the worm's forebrain. The worm's forebrain also harbors a circalunar clock entrained by nocturnal light. This monthly clock regulates maturation and persists even when circadian clock oscillations are disrupted by the inhibition of casein kinase 1δ/ε. Both circadian and circalunar clocks converge on the regulation of transcript levels. Furthermore, the circalunar clock changes the period and power of circadian behavior, although the period length of the daily transcriptional oscillations remains unaltered. We conclude that a second endogenous noncircadian clock can influence circadian clock function.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







Comment in
-
Biological clocks: riding the tides.Curr Biol. 2013 Oct 21;23(20):R921-3. doi: 10.1016/j.cub.2013.09.006. Curr Biol. 2013. PMID: 24156810 Free PMC article.
References
-
- Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A.W., Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. - PubMed
-
- Aschoff J.E. Volume 4. Plenum Press; New York: 1981. (Handbook of Behavioral Neurobiology).
Supplemental References
-
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. - PubMed
-
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. - PubMed
-
- Vinh S., Von Haeseler A. IQPNNI: moving fast through tree space and stopping in time. Mol. Biol. Evol. 2004;21:1565–1571. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

