Crystal structure of human poly(A) polymerase gamma reveals a conserved catalytic core for canonical poly(A) polymerases
- PMID: 24076191
- PMCID: PMC3878066
- DOI: 10.1016/j.jmb.2013.09.025
Crystal structure of human poly(A) polymerase gamma reveals a conserved catalytic core for canonical poly(A) polymerases
Abstract
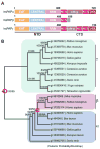
In eukaryotes, the poly(A) tail added at the 3' end of an mRNA precursor is essential for the regulation of mRNA stability and the initiation of translation. Poly(A) polymerase (PAP) is the enzyme that catalyzes the poly(A) addition reaction. Multiple isoforms of PAP have been identified in vertebrates, which originate from gene duplication, alternative splicing or post-translational modifications. The complexity of PAP isoforms suggests that they might play different roles in the cell. Phylogenetic studies indicate that vertebrate PAPs are grouped into three clades termed α, β and γ, which originated from two gene duplication events. To date, all the available PAP structures are from the PAPα clade. Here, we present the crystal structure of the first representative of the PAPγ clade, human PAPγ bound to cordycepin triphosphate (3'dATP) and Ca(2+). The structure revealed that PAPγ closely resembles its PAPα ortholog. An analysis of residue conservation reveals a conserved catalytic binding pocket, whereas residues at the surface of the polymerase are more divergent.
Keywords: 3′ end processing; C-terminal domain; CTD; MCMC; Markov Chain Monte Carlo; N-terminal domain; NLS; NTD; PAP; PEG; RNA recognition motif; RRM; mRNA processing; neo-PAP; nuclear localization signals; poly(A) polymerase; poly(A) polymerase gamma; polyadenylation; polyethylene glycol.
© 2013.
Figures

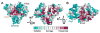
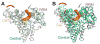
Similar articles
-
Structure of yeast poly(A) polymerase alone and in complex with 3'-dATP.Science. 2000 Aug 25;289(5483):1346-9. doi: 10.1126/science.289.5483.1346. Science. 2000. PMID: 10958780
-
Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases.Nucleic Acids Res. 2017 Sep 6;45(15):8930-8942. doi: 10.1093/nar/gkx560. Nucleic Acids Res. 2017. PMID: 28911096 Free PMC article.
-
Specific inhibition of chromatin-associated poly(A) synthesis in vitro by cordycepin 5'-triphosphate.Nature. 1977 May 12;267(5607):178-80. doi: 10.1038/267178a0. Nature. 1977. PMID: 16073440
-
Poly(A) polymerase (PAP) diversity in gene expression--star-PAP vs canonical PAP.FEBS Lett. 2014 Jun 27;588(14):2185-97. doi: 10.1016/j.febslet.2014.05.029. Epub 2014 May 27. FEBS Lett. 2014. PMID: 24873880 Free PMC article. Review.
-
Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):142-51. doi: 10.1002/wrna.16. Epub 2010 May 6. Wiley Interdiscip Rev RNA. 2010. PMID: 21956911 Review.
Cited by
-
Structure and function of pre-mRNA 5'-end capping quality control and 3'-end processing.Biochemistry. 2014 Apr 1;53(12):1882-98. doi: 10.1021/bi401715v. Epub 2014 Mar 20. Biochemistry. 2014. PMID: 24617759 Free PMC article.
-
Purification, crystallization and preliminary X-ray diffraction of the N-terminal calmodulin-like domain of the human mitochondrial ATP-Mg/Pi carrier SCaMC1.Acta Crystallogr F Struct Biol Commun. 2014 Jan;70(Pt 1):68-71. doi: 10.1107/S2053230X1303241X. Epub 2013 Dec 24. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24419621 Free PMC article.
-
Systematic analysis of the frequently amplified 2p15-p16.1 locus reveals PAPOLG as a potential proto-oncogene in follicular and transformed follicular lymphoma.Turk J Biol. 2019 Apr 5;43:124-132. doi: 10.3906/biy-1810-2. eCollection 2019. Turk J Biol. 2019. PMID: 31410080 Free PMC article.
-
Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.Nucleic Acids Res. 2020 Sep 18;48(16):9387-9405. doi: 10.1093/nar/gkaa647. Nucleic Acids Res. 2020. PMID: 32785623 Free PMC article.
-
Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.Nature. 2019 Mar;567(7747):194-199. doi: 10.1038/s41586-019-0953-5. Epub 2019 Feb 20. Nature. 2019. PMID: 30787435 Free PMC article.
References
-
- Proudfoot N, O’Sullivan J. Polyadenylation: a tail of two complexes. Curr Biol. 2002;12:R855–7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

