IFITMs restrict the replication of multiple pathogenic viruses
- PMID: 24076421
- PMCID: PMC4121887
- DOI: 10.1016/j.jmb.2013.09.024
IFITMs restrict the replication of multiple pathogenic viruses
Abstract
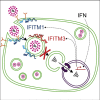
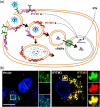
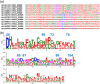
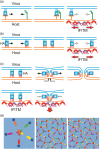
The interferon-inducible transmembrane protein (IFITM) family inhibits a growing number of pathogenic viruses, among them influenza A virus, dengue virus, hepatitis C virus, and Ebola virus. This review covers recent developments in our understanding of the IFITM's molecular determinants, potential mechanisms of action, and impact on pathogenesis.
Keywords: AS; CCHFV; CD225 family; CIL; CMEM; Crimean Congo hemorrhagic fever virus; DENV; EBOV; Ebola virus; GP; HCV; HIV-1; IAV; IFITM; IFN; JSRV; Jaagsiekte sheep retrovirus; MARV; MLV; MS; Marburg virus; Moloney leukemia virus; N-terminal domain; NTD; OSBP; RVFV; Rift Valley fever virus; SARS CoV; SeV; Sendai virus; VAPA; alanine scanning; clathrin-mediated endocytosis motif; conserved intracellular loop; dengue virus; glycoprotein; hepatitis C virus; host virus interactions; human immunodeficiency virus type 1; influenza A virus; interferon; interferon effector genes; interferon-inducible transmembrane protein; mass spectrometry; oxysterol binding protein; restriction factor; severe acute respiratory syndrome coronavirus; vesicle-associated membrane protein-A.
© 2013.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

