Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets
- PMID: 24077027
- PMCID: PMC4017624
- DOI: 10.1038/nnano.2013.194
Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets
Abstract
The spread of cancer throughout the body is driven by circulating tumour cells (CTCs). These cells detach from the primary tumour and move from the bloodstream to a new site of subsequent tumour growth. They also carry information about the primary tumour and have the potential to be valuable biomarkers for disease diagnosis and progression, and for the molecular characterization of certain biological properties of the tumour. However, the limited sensitivity and specificity of current methods for measuring and studying these cells in patient blood samples prevents the realization of their full clinical potential. The use of microfluidic devices is a promising method for isolating CTCs. However, the devices are reliant on three-dimensional structures, which limits further characterization and expansion of cells on the chip. Here we demonstrate an effective approach to isolating CTCs from blood samples of pancreatic, breast and lung cancer patients, by using functionalized graphene oxide nanosheets on a patterned gold surface. CTCs were captured with high sensitivity at a low concentration of target cells (73 ± 32.4% at 3-5 cells per ml blood).
Figures

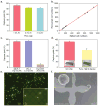

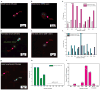
Comment in
-
New technology: nanotechnology targets cancer cells.Nat Rev Clin Oncol. 2013 Dec;10(12):667. doi: 10.1038/nrclinonc.2013.186. Epub 2013 Oct 15. Nat Rev Clin Oncol. 2013. PMID: 24129354 No abstract available.
-
Research Highlights: highlights from the latest articles in nanomedicine.Nanomedicine (Lond). 2014 Apr;9(4):385-8. doi: 10.2217/nnm.13.216. Nanomedicine (Lond). 2014. PMID: 24787437 No abstract available.
References
-
- Cristofanilli M, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. New England Journal of Medicine. 2004;351:781–791. - PubMed
-
- Willipinski-Stapelfeldt B, et al. Changes in Cytoskeletal Protein Composition Indicative of an Epithelial-Mesenchymal Transition in Human Micrometastatic and Primary Breast Carcinoma Cells. Clinical Cancer Research. 2005;11:8006–8014. - PubMed
-
- Bednarz-Knoll N, Alix-Panabières C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer and Metastasis Reviews. 2012;31:673–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

