Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru
- PMID: 24077522
- PMCID: PMC3786299
- DOI: 10.1038/srep02797
Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru
Abstract
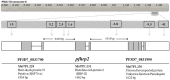
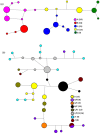
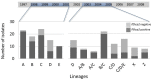
The majority of malaria rapid diagnostic tests (RDTs) detect Plasmodium falciparum histidine-rich protein 2 (PfHRP2), encoded by the pfhrp2 gene. Recently, P. falciparum isolates from Peru were found to lack pfhrp2 leading to false-negative RDT results. We hypothesized that pfhrp2-deleted parasites in Peru derived from a single genetic event. We evaluated the parasite population structure and pfhrp2 haplotype of samples collected between 1998 and 2005 using seven neutral and seven chromosome 8 microsatellite markers, respectively. Five distinct pfhrp2 haplotypes, corresponding to five neutral microsatellite-based clonal lineages, were detected in 1998-2001; pfhrp2 deletions occurred within four haplotypes. In 2003-2005, outcrossing among the parasite lineages resulted in eight population clusters that inherited the five pfhrp2 haplotypes seen previously and a new haplotype; pfhrp2 deletions occurred within four of these haplotypes. These findings indicate that the genetic origin of pfhrp2 deletion in Peru was not a single event, but likely occurred multiple times.
Figures


References
-
- Rock E. P. et al. Comparative analysis of the Plasmodium falciparum histidine-rich proteins HRP-I, HRP-II and HRP-III in malaria parasites of diverse origin. Parasitology 95 (Pt 2), 209–227 (1987). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

