Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers
- PMID: 24077665
- PMCID: PMC4226677
- DOI: 10.18632/oncotarget.1159
Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers
Abstract
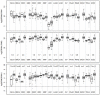
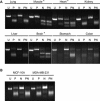
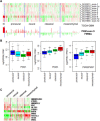
The M2 isoform of pyruvate kinase (PKM2) plays an important role in aerobic glycolysis and is a mediator of the Warburg effect in tumors. It was previously thought that tumor cells switch expression of PKM from normal tissue-expressed PKM1 to tumor-specific PKM2 via an alternative splicing mechanism. This view was challenged by a recent report demonstrating that PKM2 is already the major PKM isoform expressed in many differentiated normal tissues. Here, through analyses on sixteen tumor types using the cancer genome atlas RNA-Seq and exon array datasets, we confirmed that isoform switch from PKM1 to PKM2 occurred in glioblastomas but not in other tumor types examined. Despite lacking of isoform switches, PKM2 expression was found to be increased in all cancer types examined, and correlated strongly to poor prognosis in head and neck cancers. We further demonstrated that elevated PKM2 expression correlated well with the hypomethylation status of intron 1 of the PKM gene in multiple cancer types, suggesting epigenetic regulation by DNA methylation as a major mechanism in controlling PKM transcription in tumors. Our study suggests that isoform switch of PKM1 to PKM2 in cancers is tissue-specific and targeting PKM2 activity in tumors remains a promising approach for clinical intervention of multiple cancer types.
Figures


References
-
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. - PubMed
-
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. - PubMed
-
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. - PubMed
-
- Zhao YH, Zhou M, Liu H, Ding Y, Khong HT, Yu D, Fodstad O, Tan M. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene. 2009;28:3689–701. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

