P4-ATPases: lipid flippases in cell membranes
- PMID: 24077738
- PMCID: PMC4062807
- DOI: 10.1007/s00424-013-1363-4
P4-ATPases: lipid flippases in cell membranes
Abstract
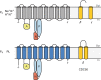
Cellular membranes, notably eukaryotic plasma membranes, are equipped with special proteins that actively translocate lipids from one leaflet to the other and thereby help generate membrane lipid asymmetry. Among these ATP-driven transporters, the P4 subfamily of P-type ATPases (P4-ATPases) comprises lipid flippases that catalyze the translocation of phospholipids from the exoplasmic to the cytosolic leaflet of cell membranes. While initially characterized as aminophospholipid translocases, recent studies of individual P4-ATPase family members from fungi, plants, and animals show that P4-ATPases differ in their substrate specificities and mediate transport of a broader range of lipid substrates, including lysophospholipids and synthetic alkylphospholipids. At the same time, the cellular processes known to be directly or indirectly affected by this class of transporters have expanded to include the regulation of membrane traffic, cytoskeletal dynamics, cell division, lipid metabolism, and lipid signaling. In this review, we will summarize the basic features of P4-ATPases and the physiological implications of their lipid transport activity in the cell.
Figures

References
-
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46(1):84–101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

