Human Polyomavirus JC monitoring and noncoding control region analysis in dynamic cohorts of individuals affected by immune-mediated diseases under treatment with biologics: an observational study
- PMID: 24079660
- PMCID: PMC3849738
- DOI: 10.1186/1743-422X-10-298
Human Polyomavirus JC monitoring and noncoding control region analysis in dynamic cohorts of individuals affected by immune-mediated diseases under treatment with biologics: an observational study
Abstract
Background: Progressive multifocal leukoencephalopathy (PML) onset, caused by Polyomavirus JC (JCPyV) in patients affected by immune-mediated diseases during biological treatment, raised concerns about the safety profile of these agents. Therefore, the aims of this study were the JCPyV reactivation monitoring and the noncoding control region (NCCR) and viral protein 1 (VP1) analysis in patients affected by different immune-mediated diseases and treated with biologics.
Methods: We performed JCPyV-specific quantitative PCR of biological samples collected at moment of recruitment (t0) and every 4 months (t1, t2, t3, t4). Subsequently, rearrangements' analysis of NCCR and VP1 was carried out. Data were analyzed using χ2 test.
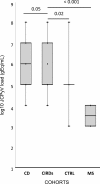
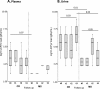
Results: Results showed that at t0 patients with chronic inflammatory rheumatic diseases presented a JCPyV load in the urine significantly higher (p≤0.05) than in patients with multiple sclerosis (MS) and Crohn's disease (CD). It can also be observed a significant association between JC viruria and JCPyV antibodies after 1 year of natalizumab (p=0.04) in MS patients. Finally, NCCR analysis showed the presence of an archetype-like sequence in all urine samples, whereas a rearranged NCCR Type IR was found in colon-rectal biopsies collected from 2 CD patients after 16 months of infliximab. Furthermore, sequences isolated from peripheral blood mononuclear cells (PBMCs) of 2 MS patients with JCPyV antibody at t0 and t3, showed a NCCR Type IIR with a duplication of a 98 bp unit and a 66 bp insert, resulting in a boxB deletion and 37 T to G transversion into the Spi-B binding site. In all patients, a prevalence of genotypes 1A and 1B, the predominant JCPyV genotypes in Europe, was observed.
Conclusions: It has been important to understand whether the specific inflammatory scenario in different immune-mediated diseases could affect JCPyV reactivation from latency, in particular from kidneys. Moreover, for a more accurate PML risk stratification, testing JC viruria seems to be useful to identify patients who harbor JCPyV but with an undetectable JCPyV-specific humoral immune response. In these patients, it may also be important to study the JCPyV NCCR rearrangement: in particular, Spi-B expression in PBMCs could play a crucial role in JCPyV replication and NCCR rearrangement.
Figures
Similar articles
-
Which is the best PML risk stratification strategy in natalizumab-treated patients affected by multiple sclerosis?Mult Scler Relat Disord. 2020 Jun;41:102008. doi: 10.1016/j.msard.2020.102008. Epub 2020 Feb 13. Mult Scler Relat Disord. 2020. PMID: 32087593
-
Human polyomavirus JC replication and non-coding control region analysis in multiple sclerosis patients under natalizumab treatment.J Neurovirol. 2015 Dec;21(6):653-65. doi: 10.1007/s13365-015-0338-y. Epub 2015 May 1. J Neurovirol. 2015. PMID: 25930159 Free PMC article.
-
Polyomavirus JC reactivation and noncoding control region sequence analysis in pediatric Crohn's disease patients treated with infliximab.J Neurovirol. 2011 Aug;17(4):303-13. doi: 10.1007/s13365-011-0036-3. Epub 2011 May 6. J Neurovirol. 2011. PMID: 21547609
-
JC polyomavirus: a short review of its biology, its association with progressive multifocal leukoencephalopathy, and the diagnostic value of different methods to manifest its activity or presence.Expert Rev Mol Diagn. 2023 Feb;23(2):143-157. doi: 10.1080/14737159.2023.2179394. Epub 2023 Feb 17. Expert Rev Mol Diagn. 2023. PMID: 36786077 Review.
-
Understanding polyomavirus CNS disease - a perspective from mouse models.FEBS J. 2022 Oct;289(19):5744-5761. doi: 10.1111/febs.16083. Epub 2021 Jul 2. FEBS J. 2022. PMID: 34145975 Free PMC article. Review.
Cited by
-
Dynamic changes of MMP-9 plasma levels correlate with JCV reactivation and immune activation in natalizumab-treated multiple sclerosis patients.Sci Rep. 2019 Jan 22;9(1):311. doi: 10.1038/s41598-018-36535-5. Sci Rep. 2019. PMID: 30670793 Free PMC article.
-
Increased Prevalence of Human Polyomavirus JC Viruria in Chronic Inflammatory Rheumatic Diseases Patients in Treatment with Anti-TNF α: A 18 Month Follow-Up Study.Front Microbiol. 2016 May 10;7:672. doi: 10.3389/fmicb.2016.00672. eCollection 2016. Front Microbiol. 2016. PMID: 27242700 Free PMC article.
-
Natalizumab Affects T-Cell Phenotype in Multiple Sclerosis: Implications for JCV Reactivation.PLoS One. 2016 Aug 3;11(8):e0160277. doi: 10.1371/journal.pone.0160277. eCollection 2016. PLoS One. 2016. PMID: 27486658 Free PMC article. Clinical Trial.
-
Immunological Markers for PML Prediction in MS Patients Treated with Natalizumab.Front Immunol. 2015 Jan 5;5:668. doi: 10.3389/fimmu.2014.00668. eCollection 2014. Front Immunol. 2015. PMID: 25601865 Free PMC article. Review.
-
Lymphocyte gene expression and JC virus noncoding control region sequences are linked with the risk of progressive multifocal leukoencephalopathy.J Virol. 2014 May;88(9):5177-83. doi: 10.1128/JVI.03221-13. Epub 2014 Feb 19. J Virol. 2014. PMID: 24554653 Free PMC article.
References
-
- Bellizzi A, Nardis C, Anzivino E, Rodìo DM, Fioriti D, Mischitelli M, Chiarini F, Pietropaolo V. Human polyomavirus JC reactivation and pathogenetic mechanisms of progressive multifocal leukoencephalopathy and cancer in the era of monoclonal antibody therapies. J Neurovirol. 2012;10:1–11. - PMC - PubMed
-
- Padgett BL, Walker DL, Zu Rhein GM, Echroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. The Lancet. 1971;10:1257–1260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

