RNA-Seq of Bacillus licheniformis: active regulatory RNA features expressed within a productive fermentation
- PMID: 24079885
- PMCID: PMC3871023
- DOI: 10.1186/1471-2164-14-667
RNA-Seq of Bacillus licheniformis: active regulatory RNA features expressed within a productive fermentation
Abstract
Background: The production of enzymes by an industrial strain requires a complex adaption of the bacterial metabolism to the conditions within the fermenter. Regulatory events within the process result in a dynamic change of the transcriptional activity of the genome. This complex network of genes is orchestrated by proteins as well as regulatory RNA elements. Here we present an RNA-Seq based study considering selected phases of an industry-oriented fermentation of Bacillus licheniformis.
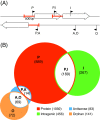
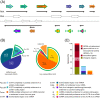
Results: A detailed analysis of 20 strand-specific RNA-Seq datasets revealed a multitude of transcriptionally active genomic regions. 3314 RNA features encoded by such active loci have been identified and sorted into ten functional classes. The identified sequences include the expected RNA features like housekeeping sRNAs, metabolic riboswitches and RNA switches well known from studies on Bacillus subtilis as well as a multitude of completely new candidates for regulatory RNAs. An unexpectedly high number of 855 RNA features are encoded antisense to annotated protein and RNA genes, in addition to 461 independently transcribed small RNAs. These antisense transcripts contain molecules with a remarkable size range variation from 38 to 6348 base pairs in length. The genome of the type strain B. licheniformis DSM13 was completely reannotated using data obtained from RNA-Seq analyses and from public databases.
Conclusion: The hereby generated data-sets represent a solid amount of knowledge on the dynamic transcriptional activities during the investigated fermentation stages. The identified regulatory elements enable research on the understanding and the optimization of crucial metabolic activities during a productive fermentation of Bacillus licheniformis strains.
Figures






References
-
- Veith B, Herzberg C, Steckel S, Feesche J, Maurer K-H, Ehrenreich P, Bäumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol. 2004;7:204–211. doi: 10.1159/000079829. - DOI - PubMed
-
- Çalık P, Takac S, Çalık G, Özdamar T. Serine alkaline protease overproduction capacity of Bacillus licheniformis. Enzyme Microb Tech. 2000;26:45–60. doi: 10.1016/S0141-0229(99)00126-X. - DOI
-
- Çalık P, Çalık G, Takaç S, Ozdamar TH. Metabolic flux analysis for serine alkaline protease fermentation by Bacillus licheniformis in a defined medium: effects of the oxygen transfer rate. Biotechnol Bioeng. 1999;64:151–167. doi: 10.1002/(SICI)1097-0290(19990720)64:2<151::AID-BIT4>3.0.CO;2-U. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

