Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response
- PMID: 24079976
- PMCID: PMC3956708
- DOI: 10.1111/cmi.12216
Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response
Abstract
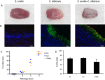
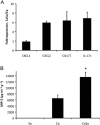
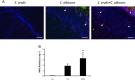
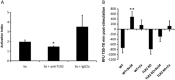
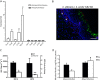
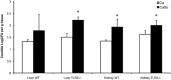
Mitis-group streptococci are ubiquitous oral commensals that can promote polybacterial biofilm virulence. Using a novel murine oral mucosal co-infection model we sought to determine for the first time whether these organisms promote the virulence of C. albicans mucosal biofilms in oropharyngeal infection and explored mechanisms of pathogenic synergy. We found that Streptococcus oralis colonization of the oral and gastrointestinal tract was augmented in the presence of C. albicans. S. oralis and C. albicans co-infection significantly augmented the frequency and size of oral thrush lesions. Importantly, S. oralis promoted deep organ dissemination of C. albicans. Whole mouse genome tongue microarray analysis showed that when compared with animals infected with one organism, the doubly infected animals had genes in the major categories of neutrophilic response/chemotaxis/inflammation significantly upregulated, indicative of an exaggerated inflammatory response. This response was dependent on TLR2 signalling since oral lesions, transcription of pro-inflammatory genes and neutrophil infiltration, were attenuated in TLR2(-/-) animals. Furthermore, S. oralis activated neutrophils in a TLR2-dependent manner in vitro. In summary, this study identifies a previously unrecognized pathogenic synergy between oral commensal bacteriaand C. albicans. This is the first report of the ability of mucosal commensal bacteria to modify the virulence of an opportunistic fungal pathogen.
© 2013 John Wiley & Sons Ltd.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

