Curli biogenesis: order out of disorder
- PMID: 24080089
- PMCID: PMC4243835
- DOI: 10.1016/j.bbamcr.2013.09.010
Curli biogenesis: order out of disorder
Abstract
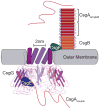
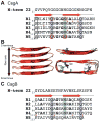
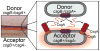
Many bacteria assemble extracellular amyloid fibers on their cell surface. Secretion of proteins across membranes and the assembly of complex macromolecular structures must be highly coordinated to avoid the accumulation of potentially toxic intracellular protein aggregates. Extracellular amyloid fiber assembly poses an even greater threat to cellular health due to the highly aggregative nature of amyloids and the inherent toxicity of amyloid assembly intermediates. Therefore, temporal and spatial control of amyloid protein secretion is paramount. The biogenesis and assembly of the extracellular bacterial amyloid curli is an ideal system for studying how bacteria cope with the many challenges of controlled and ordered amyloid assembly. Here, we review the recent progress in the curli field that has made curli biogenesis one of the best-understood functional amyloid assembly pathways. This article is part of a Special Issue entitled: Protein trafficking and secretion in bacteria. Guest Editors: Anastassios Economou and Ross Dalbey.
Keywords: Aggregate; Biofilm; Curli; Functional amyloid; Nucleation–precipitation; Type VIII secretion.
© 2013. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. - PubMed
-
- Breydo L, Wu JW, Uversky VN. Alpha-synuclein misfolding and Parkinson’s disease. Biochim Biophys Acta. 2012;1822:261–285. - PubMed
-
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

