The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula
- PMID: 24082011
- PMCID: PMC3809551
- DOI: 10.1105/tpc.113.114017
The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula
Abstract
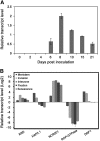
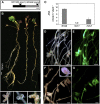
Transcription factors (TFs) are thought to regulate many aspects of nodule and symbiosis development in legumes, although few TFs have been characterized functionally. Here, we describe regulator of symbiosome differentiation (RSD) of Medicago truncatula, a member of the Cysteine-2/Histidine-2 (C2H2) family of plant TFs that is required for normal symbiosome differentiation during nodule development. RSD is expressed in a nodule-specific manner, with maximal transcript levels in the bacterial invasion zone. A tobacco (Nicotiana tabacum) retrotransposon (Tnt1) insertion rsd mutant produced nodules that were unable to fix nitrogen and that contained incompletely differentiated symbiosomes and bacteroids. RSD protein was localized to the nucleus, consistent with a role of the protein in transcriptional regulation. RSD acted as a transcriptional repressor in a heterologous yeast assay. Transcriptome analysis of an rsd mutant identified 11 genes as potential targets of RSD repression. RSD interacted physically with the promoter of one of these genes, VAMP721a, which encodes vesicle-associated membrane protein 721a. Thus, RSD may influence symbiosome development in part by repressing transcription of VAMP721a and modifying vesicle trafficking in nodule cells. This establishes RSD as a TF implicated directly in symbiosome and bacteroid differentiation and a transcriptional regulator of secretory pathway genes in plants.
Figures






Similar articles
-
Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula.Plant Cell. 2012 Sep;24(9):3838-52. doi: 10.1105/tpc.112.103267. Epub 2012 Sep 28. Plant Cell. 2012. PMID: 23023168 Free PMC article.
-
Transcription of ENOD8 in Medicago truncatula nodules directs ENOD8 esterase to developing and mature symbiosomes.Mol Plant Microbe Interact. 2008 Apr;21(4):404-10. doi: 10.1094/MPMI-21-4-0404. Mol Plant Microbe Interact. 2008. PMID: 18321186
-
A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis.Science. 2010 Feb 26;327(5969):1126-9. doi: 10.1126/science.1184096. Science. 2010. PMID: 20185723 Free PMC article.
-
Gene Expression in Nitrogen-Fixing Symbiotic Nodule Cells in Medicago truncatula and Other Nodulating Plants.Plant Cell. 2020 Jan;32(1):42-68. doi: 10.1105/tpc.19.00494. Epub 2019 Nov 11. Plant Cell. 2020. PMID: 31712407 Free PMC article. Review.
-
Symbiotic Outcome Modified by the Diversification from 7 to over 700 Nodule-Specific Cysteine-Rich Peptides.Genes (Basel). 2020 Mar 25;11(4):348. doi: 10.3390/genes11040348. Genes (Basel). 2020. PMID: 32218172 Free PMC article. Review.
Cited by
-
Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation.Plant Cell. 2020 Jan;32(1):15-41. doi: 10.1105/tpc.19.00279. Epub 2019 Oct 24. Plant Cell. 2020. PMID: 31649123 Free PMC article. Review.
-
Convergent Evolution of Endosymbiont Differentiation in Dalbergioid and Inverted Repeat-Lacking Clade Legumes Mediated by Nodule-Specific Cysteine-Rich Peptides.Plant Physiol. 2015 Oct;169(2):1254-65. doi: 10.1104/pp.15.00584. Epub 2015 Aug 18. Plant Physiol. 2015. PMID: 26286718 Free PMC article.
-
ITIS, a bioinformatics tool for accurate identification of transposon insertion sites using next-generation sequencing data.BMC Bioinformatics. 2015 Mar 5;16(1):72. doi: 10.1186/s12859-015-0507-2. BMC Bioinformatics. 2015. PMID: 25887332 Free PMC article.
-
Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis?Front Microbiol. 2014 Jun 30;5:326. doi: 10.3389/fmicb.2014.00326. eCollection 2014. Front Microbiol. 2014. PMID: 25071739 Free PMC article. Review.
-
Insight into the control of nodule immunity and senescence during Medicago truncatula symbiosis.Plant Physiol. 2023 Jan 2;191(1):729-746. doi: 10.1093/plphys/kiac505. Plant Physiol. 2023. PMID: 36305683 Free PMC article.
References
-
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513 - PubMed
-
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

