Guidelines for the design, analysis and interpretation of 'omics' data: focus on human endometrium
- PMID: 24082038
- PMCID: PMC3845681
- DOI: 10.1093/humupd/dmt048
Guidelines for the design, analysis and interpretation of 'omics' data: focus on human endometrium
Abstract
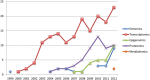
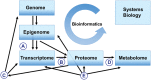
Background: 'Omics' high-throughput analyses, including genomics, epigenomics, transcriptomics, proteomics and metabolomics, are widely applied in human endometrial studies. Analysis of endometrial transcriptome patterns in physiological and pathophysiological conditions has been to date the most commonly applied 'omics' technique in human endometrium. As the technologies improve, proteomics holds the next big promise for this field. The 'omics' technologies have undoubtedly advanced our knowledge of human endometrium in relation to fertility and different diseases. Nevertheless, the challenges arising from the vast amount of data generated and the broad variation of 'omics' profiling according to different environments and stimuli make it difficult to assess the validity, reproducibility and interpretation of such 'omics' data. With the expansion of 'omics' analyses in the study of the endometrium, there is a growing need to develop guidelines for the design of studies, and the analysis and interpretation of 'omics' data.
Methods: Systematic review of the literature in PubMed, and references from relevant articles were investigated up to March 2013.
Results: The current review aims to provide guidelines for future 'omics' studies on human endometrium, together with a summary of the status and trends, promise and shortcomings in the high-throughput technologies. In addition, the approaches presented here can be adapted to other areas of high-throughput 'omics' studies.
Conclusion: A highly rigorous approach to future studies, based on the guidelines provided here, is a prerequisite for obtaining data on biological systems which can be shared among researchers worldwide and will ultimately be of clinical benefit.
Keywords: endometrium; epigenomics; genomics; metabolomics; proteomics.
Figures

References
-
- Adachi S, Tajima A, Quan J, Haino K, Yoshihara K, Masuzaki H, Katabuchi H, Ikuma K, Suginami H, Nishida N, et al. Meta-analysis of genome-wide association scans for genetic susceptibility to endometriosis in Japanese population. J Hum Genet. 2010;55:816–821. - PubMed
-
- Aghajanova L, Simón C, Horcajadas JA. Are favorite molecules of endometrial receptivity still in favor? Expert Rev Obstet Gynecol. 2008b;3:487–501.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

