Neurexins
- PMID: 24083347
- PMCID: PMC4056431
- DOI: 10.1186/gb-2013-14-9-213
Neurexins
Abstract
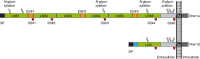
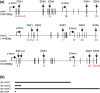
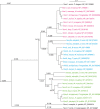
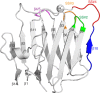
The neurexin family of cell adhesion proteins consists of three members in vertebrates and has homologs in several invertebrate species. In mammals, each neurexin gene encodes an α-neurexin in which the extracellular portion is long, and a β-neurexin in which the extracellular portion is short. As a result of alternative splicing, both major isoforms can be transcribed in many variants, contributing to distinct structural domains and variability. Neurexins act predominantly at the presynaptic terminal in neurons and play essential roles in neurotransmission and differentiation of synapses. Some of these functions require the formation of trans-synaptic complexes with postsynaptic proteins such as neuroligins, LRRTM proteins or cerebellin. In addition, rare mutations and copy-number variations of human neurexin genes have been linked to autism and schizophrenia, indicating that impairments of synaptic function sustained by neurexins and their binding partners maybe relevant to the pathomechanism of these debilitating diseases.
Figures


References
-
- Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. - PubMed
-
- Ushkaryov YA, Hata Y, Ichtchenko K, Moomaw C, Afendis S, Slaughter CA, Südhof TC. Conserved domain structure of beta-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 1994;14:11987–11992. - PubMed
-
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;14:50–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

