Microbial community composition of Tirez lagoon (Spain), a highly sulfated athalassohaline environment
- PMID: 24083554
- PMCID: PMC3852488
- DOI: 10.1186/2046-9063-9-19
Microbial community composition of Tirez lagoon (Spain), a highly sulfated athalassohaline environment
Abstract
Background: The aim was to study the seasonal microbial diversity variations of an athalassohaline environment with a high concentration of sulfates in Tirez lagoon (La Mancha, Spain). Despite the interest in these types of environments there is scarce information about their microbial ecology, especially on their anoxic sediments.
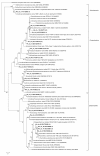
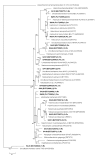
Results: We report the seasonal microbial diversity of the water column and the sediments of a highly sulfated lagoon using both molecular and conventional microbiological methods. Algae and Cyanobacteria were the main photosynthetic primary producers detected in the ecosystem in the rainy season. Also dinoflagelates and filamentous fungi were identified in the brines. The highest phylotype abundance in water and sediments corresponded to members of the bacterial phylum Proteobacteria, mainly of the Gamma- and Alphaproteobacteria classes. Firmicutes and Actinobacteria were isolated and identified in Tirez brines and sediment samples. Halophilic sulfate reducing Deltaproteobacteria were also detected (Desulfohalobium).
Conclusions: Important differences have been found in the microbial diversity present in the Tirez water column and the sediments between the wet and dry seasons. Also the Tirez lagoon showed a high richness of the bacterial Alpha- and Deltaproteobacteria, Bacteroidetes, Firmicutes, Actinobacteria and for the archaeal Euryarchaeota.
Figures



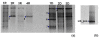
References
-
- Sorokin DY, Kuenen JG, Muyzer G. The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Front Microbiol. 2011;2:44. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3128939/ - PMC - PubMed
-
- vd Wielen PWJJ, Bolhuis H, Borin S, Daffonchio D, Corselli C, Giuliano L, D’Auria G, de Lange GJ, Huebner A, Varnavas SP, Thomson J, Tamburini C, Marty D, McGenity TJ, Timmis KN. BioDeep Scientific Party. The Enigma of Prokaryotic Life in Deep Hypersaline Anoxic Basins. Science. 2005;307:121–123. doi: 10.1126/science.1103569. http://www.sciencemag.org/cgi/content/abstract/307/5706/121. - DOI - PubMed
-
- Porter D, Roychoudhury AN, Cowan D. Dissimilatory sulfate reduction in hypersaline coastal pans: Activity across a salinity gradient. Geochim Cosmochim Acta. 2007;71:5102–5116. doi: 10.1016/j.gca.2007.08.023. http://www.sciencedirect.com/science/article/pii/S0016703707004954. - DOI
-
- Borin S, Brusetti L, Mapelli F, D’Auria G, Brusa T, Marzorati M, Rizzi A, Yakimov M, Marty D, De Lange GJ. et al. Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc Natl Acad Sci U S A. 2009;106:9151–9156. doi: 10.1073/pnas.0811984106. http://www.pnas.org/content/106/23/9151.short. - DOI - PMC - PubMed
-
- Sorokin DY, Zacharova EE, Pimenov NV, Tourova TP, Panteleeva AN, Muyzer G. Sulfidogenesis in hypersaline chloride-sulfate lakes of Kulunda Steppe (Altai, Russia) FEMS Microbiol Ecol. 2012;79:445–453. doi: 10.1111/j.1574-6941.2011.01228.x. http://www.ncbi.nlm.nih.gov/pubmed/22092787. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

