The immediate upstream region of the 5'-UTR from the AUG start codon has a pronounced effect on the translational efficiency in Arabidopsis thaliana
- PMID: 24084084
- PMCID: PMC3874180
- DOI: 10.1093/nar/gkt864
The immediate upstream region of the 5'-UTR from the AUG start codon has a pronounced effect on the translational efficiency in Arabidopsis thaliana
Abstract
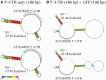
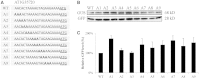
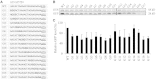
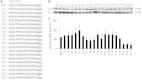
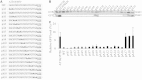
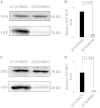
The nucleotide sequence around the translational initiation site is an important cis-acting element for post-transcriptional regulation. However, it has not been fully understood how the sequence context at the 5'-untranslated region (5'-UTR) affects the translational efficiency of individual mRNAs. In this study, we provide evidence that the 5'-UTRs of Arabidopsis genes showing a great difference in the nucleotide sequence vary greatly in translational efficiency with more than a 200-fold difference. Of the four types of nucleotides, the A residue was the most favourable nucleotide from positions -1 to -21 of the 5'-UTRs in Arabidopsis genes. In particular, the A residue in the 5'-UTR from positions -1 to -5 was required for a high-level translational efficiency. In contrast, the T residue in the 5'-UTR from positions -1 to -5 was the least favourable nucleotide in translational efficiency. Furthermore, the effect of the sequence context in the -1 to -21 region of the 5'-UTR was conserved in different plant species. Based on these observations, we propose that the sequence context immediately upstream of the AUG initiation codon plays a crucial role in determining the translational efficiency of plant genes.
Figures





References
-
- van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell. Biol. 1999;31:87–106. - PubMed
-
- Jansen RP. mRNA localization: message on the move. Nat. Rev. Mol. Cell. Biol. 2001;2:247–256. - PubMed
-
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

