The genomic distribution of sex-biased genes in drosophila serrata: X chromosome demasculinization, feminization, and hyperexpression in both sexes
- PMID: 24084777
- PMCID: PMC3814203
- DOI: 10.1093/gbe/evt145
The genomic distribution of sex-biased genes in drosophila serrata: X chromosome demasculinization, feminization, and hyperexpression in both sexes
Abstract
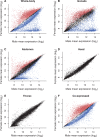
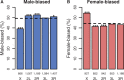
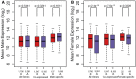
The chromosomal distribution of genes with sex-biased expression is often nonrandom, and in species with XY sex chromosome systems, it is common to observe a deficit of X-linked male-biased genes and an excess of X-linked female-biased genes. One explanation for this pattern is that sex-specific selection has shaped the gene content of the X. Alternatively, the deficit of male-biased and excess of female-biased genes could be an artifact of differences between the sexes in the global expression level of their X chromosome(s), perhaps brought about by a lack of dosage compensation in males and hyperexpression in females. In the montium fruit fly, Drosophila serrata, both these explanations can account for a deficit of male-biased and excess of female-biased X-linked genes. Using genome-wide expression data from multiple male and female tissues (n = 176 hybridizations), we found that testis- and accessory gland-specific genes are underrepresented whereas female ovary-specific genes are overrepresented on the X chromosome, suggesting that X-linkage is disfavored for male function genes but favored for female function genes. However, genes with such sex-specific functions did not fully account for the deficit of male-biased and excess of female-biased X-linked genes. We did, however, observe sex differences in the global expression level of the X chromosome and autosomes. Surprisingly, and in contrast to other species where a lack of dosage compensation in males is responsible, we found that hyperexpression of X-linked genes in both sexes leads to this imbalance in D. serrata. Our results highlight how common genomic distributions of sex-biased genes, even among closely related species, may arise via quite different evolutionary processes.
Keywords: Drosophila; X chromosome; dosage compensation; sex-biased gene expression.
Figures

References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

