Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling
- PMID: 24084807
- PMCID: PMC3817400
- DOI: 10.1038/msb.2013.50
Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling
Abstract
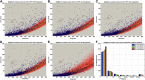
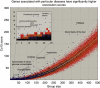
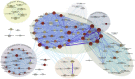
Genes with common profiles of the presence and absence in disparate genomes tend to function in the same pathway. By mapping all human genes into about 1000 clusters of genes with similar patterns of conservation across eukaryotic phylogeny, we determined that sets of genes associated with particular diseases have similar phylogenetic profiles. By focusing on those human phylogenetic gene clusters that significantly overlap some of the thousands of human gene sets defined by their coexpression or annotation to pathways or other molecular attributes, we reveal the evolutionary map that connects molecular pathways and human diseases. The other genes in the phylogenetic clusters enriched for particular known disease genes or molecular pathways identify candidate genes for roles in those same disorders and pathways. Focusing on proteins coevolved with the microphthalmia-associated transcription factor (MITF), we identified the Notch pathway suppressor of hairless (RBP-Jk/SuH) transcription factor, and showed that RBP-Jk functions as an MITF cofactor.
Figures





References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular Biology of the Cell, 4th edn. Garland Science, New York, USA
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Aubin-Houzelstein G, Djian-Zaouche J, Bernex F, Gadin S, Delmas V, Larue L, Panthier JJ (2008) Melanoblasts' proper location and timed differentiation depend on Notch/RBP-J signaling in postnatal hair follicles. J Invest Dermatol 128: 2686–2695 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

