Comprehensive identification of mutational cancer driver genes across 12 tumor types
- PMID: 24084849
- PMCID: PMC3788361
- DOI: 10.1038/srep02650
Comprehensive identification of mutational cancer driver genes across 12 tumor types
Abstract
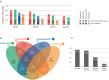
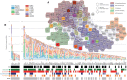
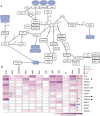
With the ability to fully sequence tumor genomes/exomes, the quest for cancer driver genes can now be undertaken in an unbiased manner. However, obtaining a complete catalog of cancer genes is difficult due to the heterogeneous molecular nature of the disease and the limitations of available computational methods. Here we show that the combination of complementary methods allows identifying a comprehensive and reliable list of cancer driver genes. We provide a list of 291 high-confidence cancer driver genes acting on 3,205 tumors from 12 different cancer types. Among those genes, some have not been previously identified as cancer drivers and 16 have clear preference to sustain mutations in one specific tumor type. The novel driver candidates complement our current picture of the emergence of these diseases. In summary, the catalog of driver genes and the methodology presented here open new avenues to better understand the mechanisms of tumorigenesis.
Figures


References
-
- Reddy E. P., Reynolds R. K., Santos E. & Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature 300, 149–152 (1982). - PubMed
-
- Tabin C. J. et al. Mechanism of activation of a human oncogene. Nature 300, 143–149 (1982). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

